Structural insights into cell cycle control by essential GTPase Era
- PMID: 28132488
- PMCID: PMC6622462
Structural insights into cell cycle control by essential GTPase Era
Abstract
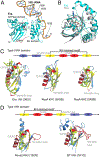
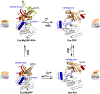
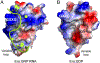
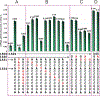
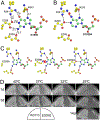
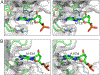
Era (Escherichia coli Ras-like protein), essential for bacterial cell viability, is composed of an N-terminal GTPase domain and a C-terminal KH domain. In bacteria, it is required for the processing of 16S ribosomal RNA (rRNA) and maturation of 30S (small) ribosomal subunit. Era recognizes 10 nucleotides (1530GAUCACCUCC1539) near the 3' end of 16S rRNA and interacts with helix 45 (h45, nucleotides 1506-1529). GTP binding enables Era to bind RNA, RNA binding stimulates Era's GTP-hydrolyzing activity, and GTP hydrolysis releases Era from matured 30S ribosomal subunit. As such, Era controls cell growth rate via regulating the maturation of the 30S ribosomal subunit. Ribosomes manufacture proteins in all living organisms. The GAUCA sequence and h45 are highly conserved in all three kingdoms of life. Homologues of Era are present in eukaryotic cells. Hence, the mechanism of bacterial Era action also sheds light on the cell cycle control of eukaryotes.
Białko Era (ang. Escherichia coli Ras-like protein), niezbędne dla funkcjonowania komórki bakteryjnej, składa się z N-końcowej domeny o aktywności GTPazy oraz C-końcowej domeny KH. W komórce bakteryjnej białko to uczestniczy w modyfikacji rybosomalnego RNA (rRNA) 16S oraz dojrzewaniu podjednostki 30S (małej) rybosomu. Era rozpoznaje sekwencję 10 nukleotydów (1530GAUCACCUCC1539) w pobliżu końca 3' rRNA 16S i oddziałuje z helisą 45 (h45, nukleotydy 1506-1529). Związanie GTP umożliwia związanie przez białko Era cząsteczki RNA, co zwiększa jego aktywność hydrolizy GTP. W wyniku hydrolizy GTP białko Era zostaje z kolei uwolnione od dojrzałej podjednostki 30S rybosomu. W ten sposób Era kontroluje szybkość wzrostu komórki poprzez regulację dojrzewania podjednostek 30S rybosomów. Jak wiadomo, rybosomy są miejscem produkcji białek w każdej komórce. Sekwencja GAUCA i struktura h45 są zachowane w ewolucji we wszystkich trzech królestwach organizmów żywych. Homologi białka Era wykryto również w komórkach eukariotycznych. Z tego względu zrozumienie mechanizmu działania bakteryjnego białka Era dostarcza ważnych wskazówek dotyczących kontroli cyklu komórkowego także w komórkach Eukaryota.
Keywords: 16S rRNA; 30S ribosomal subunit; cell cycle control; ribosome biogenesis.
Figures

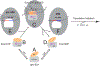
Similar articles
-
The Era GTPase recognizes the GAUCACCUCC sequence and binds helix 45 near the 3' end of 16S rRNA.Proc Natl Acad Sci U S A. 2011 Jun 21;108(25):10156-61. doi: 10.1073/pnas.1017679108. Epub 2011 Jun 6. Proc Natl Acad Sci U S A. 2011. PMID: 21646538 Free PMC article.
-
Suppression of defective ribosome assembly in a rbfA deletion mutant by overexpression of Era, an essential GTPase in Escherichia coli.Mol Microbiol. 2003 May;48(4):1005-16. doi: 10.1046/j.1365-2958.2003.03475.x. Mol Microbiol. 2003. PMID: 12753192
-
Role of Era in assembly and homeostasis of the ribosomal small subunit.Nucleic Acids Res. 2019 Sep 5;47(15):8301-8317. doi: 10.1093/nar/gkz571. Nucleic Acids Res. 2019. PMID: 31265110 Free PMC article.
-
Interaction of Era with the 30S ribosomal subunit implications for 30S subunit assembly.Mol Cell. 2005 Apr 29;18(3):319-29. doi: 10.1016/j.molcel.2005.03.028. Mol Cell. 2005. PMID: 15866174
-
RNA-protein interactions in 30S ribosomal subunits: folding and function of 16S rRNA.Science. 1989 May 19;244(4906):783-90. doi: 10.1126/science.2658053. Science. 1989. PMID: 2658053 Review.
Cited by
-
Reformulation of an extant ATPase active site to mimic ancestral GTPase activity reveals a nucleotide base requirement for function.Elife. 2021 Mar 11;10:e65845. doi: 10.7554/eLife.65845. Elife. 2021. PMID: 33704064 Free PMC article.
-
GTPase Era at the heart of ribosome assembly.Front Mol Biosci. 2023 Oct 4;10:1263433. doi: 10.3389/fmolb.2023.1263433. eCollection 2023. Front Mol Biosci. 2023. PMID: 37860580 Free PMC article. Review.
-
WSL6 encoding an Era-type GTP-binding protein is essential for chloroplast development in rice.Plant Mol Biol. 2019 Aug;100(6):635-645. doi: 10.1007/s11103-019-00885-z. Epub 2019 May 30. Plant Mol Biol. 2019. PMID: 31147815
-
The Impact of the Stringent Response on TRAFAC GTPases and Prokaryotic Ribosome Assembly.Cells. 2019 Oct 24;8(11):1313. doi: 10.3390/cells8111313. Cells. 2019. PMID: 31653044 Free PMC article. Review.
-
Contribution of YthA, a PspC Family Transcriptional Regulator of Lactococcus lactis F44 Acid Tolerance and Nisin Yield: a Transcriptomic Approach.Appl Environ Microbiol. 2018 Mar 1;84(6):e02483-17. doi: 10.1128/AEM.02483-17. Print 2018 Mar 15. Appl Environ Microbiol. 2018. PMID: 29305506 Free PMC article.
References
-
- March PE, Lerner CG, Ahnn J, Cui X, Inouye M (1988) The Escherichia coli Ras-like protein (Era) has GTPase activity and is essential for cell growth. Oncogene 2: 539–544 - PubMed
-
- Zuber M, Hoover TA, Dertzbaugh MT, Court DL (1997) A Francisella tularensis DNA clone complements Escherichia coli defective for the production of Era, an essential Ras-like GTP-binding protein. Gene 189: 31–34 - PubMed
-
- Eltsov M, Zuber B (2006) Transmission electron microscopy of the bacterial nucleoid. J Struct Biol 156: 246–254 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
