The ER-Mitochondria Tethering Complex VAPB-PTPIP51 Regulates Autophagy
- PMID: 28132811
- PMCID: PMC5300905
- DOI: 10.1016/j.cub.2016.12.038
The ER-Mitochondria Tethering Complex VAPB-PTPIP51 Regulates Autophagy
Abstract
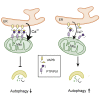
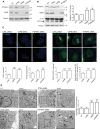
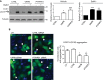
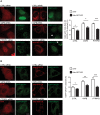
Mitochondria form close physical associations with the endoplasmic reticulum (ER) that regulate a number of physiological functions. One mechanism by which regions of ER are recruited to mitochondria involves binding of the ER protein VAPB to the mitochondrial protein PTPIP51, which act as scaffolds to tether the two organelles. Here, we show that the VAPB-PTPIP51 tethers regulate autophagy. We demonstrate that overexpression of VAPB or PTPIP51 to tighten ER-mitochondria contacts impairs, whereas small interfering RNA (siRNA)-mediated loss of VAPB or PTPIP51 to loosen contacts stimulates, autophagosome formation. Moreover, we show that expression of a synthetic linker protein that artificially tethers ER and mitochondria also reduces autophagosome formation, and that this artificial tether rescues the effects of siRNA loss of VAPB or PTPIP51 on autophagy. Thus, these effects of VAPB and PTPIP51 manipulation on autophagy are a consequence of their ER-mitochondria tethering function. Interestingly, we discovered that tightening of ER-mitochondria contacts by overexpression of VAPB or PTPIP51 impairs rapamycin- and torin 1-induced, but not starvation-induced, autophagy. This suggests that the regulation of autophagy by ER-mitochondria signaling is at least partly dependent upon the nature of the autophagic stimulus. Finally, we demonstrate that the mechanism by which the VAPB-PTPIP51 tethers regulate autophagy involves their role in mediating delivery of Ca2+ to mitochondria from ER stores. Thus, our findings reveal a new molecular mechanism for regulating autophagy.
Keywords: Alzheimer’s disease; MAM; PTPIP51; Parkinson’s disease; VAPB; amyotrophic lateral sclerosis; autophagy; calcium; endoplasmic reticulum; mitochondria.
Copyright © 2017 The Author(s). Published by Elsevier Ltd.. All rights reserved.
Figures




Comment in
-
Organelle dynamics: Connections, connections, connections.Nat Rev Mol Cell Biol. 2017 Feb 21;18(3):139. doi: 10.1038/nrm.2017.14. Nat Rev Mol Cell Biol. 2017. PMID: 28220048 No abstract available.
References
-
- Mizushima N. Autophagy: process and function. Genes Dev. 2007;21:2861–2873. - PubMed
-
- Lamb C.A., Yoshimori T., Tooze S.A. The autophagosome: origins unknown, biogenesis complex. Nat. Rev. Mol. Cell Biol. 2013;14:759–774. - PubMed
-
- Westrate L.M., Lee J.E., Prinz W.A., Voeltz G.K. Form follows function: the importance of endoplasmic reticulum shape. Annu. Rev. Biochem. 2015;84:791–811. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

