Systematic Mapping of RNA-Chromatin Interactions In Vivo
- PMID: 28132817
- PMCID: PMC5319903
- DOI: 10.1016/j.cub.2017.01.011
Systematic Mapping of RNA-Chromatin Interactions In Vivo
Erratum in
-
Systematic Mapping of RNA-Chromatin Interactions In Vivo.Curr Biol. 2017 Feb 20;27(4):610-612. doi: 10.1016/j.cub.2017.01.068. Curr Biol. 2017. PMID: 28222283 No abstract available.
Abstract
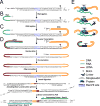
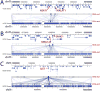
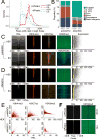
RNA molecules can attach to chromatin. It remains difficult to know what RNAs are associated with chromatin and where the genomic target loci of these RNAs are. Here, we present MARGI (mapping RNA-genome interactions), a technology to massively reveal native RNA-chromatin interactions from unperturbed cells. The gist of this technology is to ligate chromatin-associated RNAs (caRNAs) with their target genomic sequences by proximity ligation, forming RNA-DNA chimeric sequences, which are converted to a sequencing library for paired-end sequencing. Using MARGI, we produced RNA-genome interaction maps for human embryonic stem cells (ESCs) and human embryonic kidney (HEK) cells. MARGI revealed hundreds of caRNAs, including previously known XIST, SNHG1, NEAT1, and MALAT1, as well as each caRNA's genomic interaction loci. Using a cross-species experiment, we estimated that approximately 2.2% of MARGI-identified interactions were false positives. In ESCs and HEK cells, the RNA ends of more than 5% of MARGI read pairs were mapped to distal or inter-chromosomal locations as compared to the locations of their corresponding DNA ends. The majority of transcription start sites are associated with distal or inter-chromosomal caRNAs. Chromatin-immunoprecipitation-sequencing (ChIP-seq)-reported H3K27ac and H3K4me3 levels are positively correlated, while H3K9me3 is negatively correlated, with MARGI-reported RNA attachment levels. The MARGI technology should facilitate revealing novel RNA functions and their genomic target regions.
Keywords: RNA; RNA-chromatin interactions; chromatin; chromatin-associated RNA; proximity ligation.
Copyright © 2017 Elsevier Ltd. All rights reserved.
Figures

References
-
- Yin S, Ho CK, Shuman S. Structure-function analysis of T4 RNA ligase 2. The Journal of biological chemistry. 2003;278:17601–17608. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

