Nicotinamide Ameliorates Disease Phenotypes in a Human iPSC Model of Age-Related Macular Degeneration
- PMID: 28132833
- PMCID: PMC5419856
- DOI: 10.1016/j.stem.2016.12.015
Nicotinamide Ameliorates Disease Phenotypes in a Human iPSC Model of Age-Related Macular Degeneration
Abstract
Age-related macular degeneration (AMD) affects the retinal pigment epithelium (RPE), a cell monolayer essential for photoreceptor survival, and is the leading cause of vision loss in the elderly. There are no disease-altering therapies for dry AMD, which is characterized by accumulation of subretinal drusen deposits and complement-driven inflammation. We report the derivation of human-induced pluripotent stem cells (hiPSCs) from patients with diagnosed AMD, including two donors with the rare ARMS2/HTRA1 homozygous genotype. The hiPSC-derived RPE cells produce several AMD/drusen-related proteins, and those from the AMD donors show significantly increased complement and inflammatory factors, which are most exaggerated in the ARMS2/HTRA1 lines. Using a panel of AMD biomarkers and candidate drug screening, combined with transcriptome analysis, we discover that nicotinamide (NAM) ameliorated disease-related phenotypes by inhibiting drusen proteins and inflammatory and complement factors while upregulating nucleosome, ribosome, and chromatin-modifying genes. Thus, targeting NAM-regulated pathways is a promising avenue for developing therapeutics to combat AMD.
Keywords: age-related macular degeneration; aging; complement; drusen; human induced pluripotent stem cells; nicotinamide; retina; retinal pigment epithelium.
Copyright © 2017 Elsevier Inc. All rights reserved.
Figures
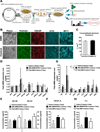



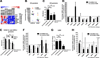
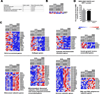
Comment in
-
Eyeing the Fountain of Youth.Cell Stem Cell. 2017 May 4;20(5):583-584. doi: 10.1016/j.stem.2017.04.007. Cell Stem Cell. 2017. PMID: 28475881
-
Nicotinamide, iRPE-in-a dish, and age-related macular degeneration therapy development.Stem Cell Investig. 2017 Sep 29;4:81. doi: 10.21037/sci.2017.09.05. eCollection 2017. Stem Cell Investig. 2017. PMID: 29057253 Free PMC article. No abstract available.
-
Nicotinamide: a novel treatment for age-related macular degeneration?Stem Cell Investig. 2017 Oct 27;4:86. doi: 10.21037/sci.2017.10.01. eCollection 2017. Stem Cell Investig. 2017. PMID: 29167807 Free PMC article. No abstract available.
-
Human induced pluripotent stem cells illuminate pathways and novel treatment targets for age-related macular degeneration.Stem Cell Investig. 2017 Nov 17;4:92. doi: 10.21037/sci.2017.10.07. eCollection 2017. Stem Cell Investig. 2017. PMID: 29270418 Free PMC article. No abstract available.
References
-
- An E, Lu X, Flippin J, Devaney JM, Halligan B, Hoffman EP, Strunnikova N, Csaky K, Hathout Y. Secreted proteome profiling in human RPE cell cultures derived from donors with age related macular degeneration and age matched healthy donors. Journal of proteome research. 2006;5:2599–2610. - PubMed
-
- Blenkinsop TA, Saini JS, Maminishkis A, Bharti K, Wan Q, Banzon T, Lotfi M, Davis J, Singh D, Rizzolo LJ, et al. Human Adult Retinal Pigment Epithelial Stem Cell-Derived RPE Monolayers Exhibit Key Physiological Characteristics of Native Tissue. Investigative ophthalmology & visual science. 2015;56:7085–7099. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

