Vernalization-Triggered Intragenic Chromatin Loop Formation by Long Noncoding RNAs
- PMID: 28132848
- PMCID: PMC5303624
- DOI: 10.1016/j.devcel.2016.12.021
Vernalization-Triggered Intragenic Chromatin Loop Formation by Long Noncoding RNAs
Abstract
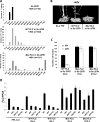
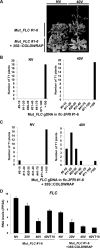
Long noncoding RNAs (lncRNAs) affect gene regulation through structural and regulatory interactions with associated proteins. The Polycomb complex often binds to lncRNAs in eukaryotes, and an lncRNA, COLDAIR, associates with Polycomb to mediate silencing of the floral repressor FLOWERING LOCUS C (FLC) during the process of vernalization in Arabidopsis. Here, we identified an additional Polycomb-binding lncRNA, COLDWRAP. COLDWRAP is derived from the repressed promoter of FLC and is necessary for the establishment of the stable repressed state of FLC by vernalization. Both COLDAIR and COLDWRAP are required to form a repressive intragenic chromatin loop at the FLC locus by vernalization. Our results indicate that vernalization-mediated Polycomb silencing is coordinated by lncRNAs in a cooperative manner to form a stable repressive chromatin structure.
Keywords: chromatin loop; flowering; long noncoding RNAs; polycomb repressive complex 2; vernalization.
Copyright © 2017 Elsevier Inc. All rights reserved.
Figures
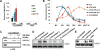



References
-
- Andres F, Coupland G. The genetic basis of flowering responses to season al cues. Nat Rev Genet. 2012;13:627–639. - PubMed
-
- Angel A, Song J, Dean C, Howard M. A Polycomb-based switch underlying quantitative epigenetic memory. Nature. 2011;476:105–108. - PubMed
-
- Ariel F, Jegu T, Latrasse D, Romero-Barrios N, Christ A, Benhamed M, Crespi M. Noncoding transcription by alternative RNA polymerases dynamically regulates an auxin-driven chromatin loop. Mol Cell. 2014;55:383–396. - PubMed
-
- Bastow R, Mylne JS, Lister C, Lippman Z, Martienssen RA, Dean C. Vernalization requires epigenetic silencing of FLC by histone methylation. Nature. 2004;427:164–167. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

