Capitalizing on disaster: Establishing chromatin specificity behind the replication fork
- PMID: 28133760
- PMCID: PMC5513704
- DOI: 10.1002/bies.201600150
Capitalizing on disaster: Establishing chromatin specificity behind the replication fork
Abstract
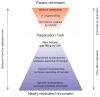
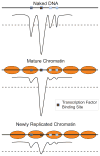
Eukaryotic genomes are packaged into nucleosomal chromatin, and genomic activity requires the precise localization of transcription factors, histone modifications and nucleosomes. Classic work described the progressive reassembly and maturation of bulk chromatin behind replication forks. More recent proteomics has detailed the molecular machines that accompany the replicative polymerase to promote rapid histone deposition onto the newly replicated DNA. However, localized chromatin features are transiently obliterated by DNA replication every S phase of the cell cycle. Genomic strategies now observe the rebuilding of locus-specific chromatin features, and reveal surprising delays in transcription factor binding behind replication forks. This implies that transient chromatin disorganization during replication is a central juncture for targeted transcription factor binding within genomes. We propose that transient occlusion of regulatory elements by disorganized nucleosomes during chromatin maturation enforces specificity of factor binding.
Keywords: chromatin; epigenetics.
© 2017 WILEY Periodicals, Inc.
Figures

References
-
- Smith PA, Jackson V, Chalkley R. Two-stage maturation process for newly replicated chromatin. Biochemistry. 1984;23:1576–81. - PubMed
-
- Xu M, Long C, Chen X, Huang C, et al. Partitioning of histone H3–H4 tetramers during DNA replication-dependent chromatin assembly. Science. 2010;328:94–8. - PubMed
-
- Jackson V, Chalkley R. Histone segregation on replicating chromatin. Biochemistry. 1985;24:6930–8. - PubMed
-
- Annunziato AT, Seale RL. Histone deacetylation is required for the maturation of newly replicated chromatin. J Biol Chem. 1983;258:12675–84. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

