Boundaries of loop domains (insulators): Determinants of chromosome form and function in multicellular eukaryotes
- PMID: 28133765
- PMCID: PMC5536339
- DOI: 10.1002/bies.201600233
Boundaries of loop domains (insulators): Determinants of chromosome form and function in multicellular eukaryotes
Abstract
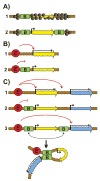
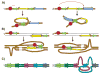
Chromosomes in multicellular animals are subdivided into a series of looped domains. In addition to being the underlying principle for organizing the chromatin fiber, looping is critical for processes ranging from gene regulation to recombination and repair. The subdivision of chromosomes into looped domains depends upon a special class of architectural elements called boundaries or insulators. These elements are distributed throughout the genome and are ubiquitous building blocks of chromosomes. In this review, we focus on features of boundaries that are critical in determining the topology of the looped domains and their genetic properties. We highlight the properties of fly boundaries that are likely to have an important bearing on the organization of looped domains in vertebrates, and discuss the functional consequences of the observed similarities and differences.
Keywords: TADs; chromosome architecture; gene regulation; insulators; loop topology; looped domain.
© 2017 WILEY Periodicals, Inc.
Figures



References
-
- Bickmore WA. The spatial organization of the human genome. Annu Review Genomics Hum Genet. 2013;14:67–84. - PubMed
-
- Ciabrelli F, Cavalli G. Chromatin-driven behavior of topologically associating domains. J Mol Biol. 2015;427:608–25. - PubMed
-
- Cremer T, Cremer M, Hubner B, Strickfaden H, et al. The 4D nucleome: Evidence for a dynamic nuclear landscape based on co-aligned active and inactive nuclear compartments. FEBS Lett. 2015;589:2931–43. - PubMed
-
- Misteli T. Beyond the sequence: cellular organization of genome function. Cell. 2007;128:787–800. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

