Polyvalent vaccine approaches to combat HIV-1 diversity
- PMID: 28133800
- PMCID: PMC5362114
- DOI: 10.1111/imr.12516
Polyvalent vaccine approaches to combat HIV-1 diversity
Abstract
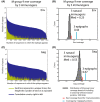
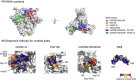
A key unresolved challenge for developing an effective HIV-1 vaccine is the discovery of strategies to elicit immune responses that are able to cross-protect against a significant fraction of the diverse viruses that are circulating worldwide. Here, we summarize some of the immunological implications of HIV-1 diversity, and outline the rationale behind several polyvalent vaccine design strategies that are currently under evaluation. Vaccine-elicited T-cell responses, which contribute to the control of HIV-1 in natural infections, are currently being considered in both prevention and treatment settings. Approaches now in preclinical and human trials include full proteins in novel vectors, concatenated conserved protein regions, and polyvalent strategies that improve coverage of epitope diversity and enhance the cross-reactivity of responses. While many barriers to vaccine induction of broadly neutralizing antibody (bNAb) responses remain, epitope diversification has emerged as both a challenge and an opportunity. Recent longitudinal studies have traced the emergence of bNAbs in HIV-1 infection, inspiring novel approaches to recapitulate and accelerate the events that give rise to potent bNAb in vivo. In this review, we have selected two such lineage-based design strategies to illustrate how such in-depth analysis can offer conceptual improvements that may bring us closer to an effective vaccine.
Keywords: AIDS; B cells; antibodies; antigens/peptides/epitopes; vaccination; viral.
Published 2017. This article is a U.S. Government work and is in the public domain in the USA. Immunological Reviews published by John Wiley & Sons Ltd.
Figures




References
-
- Gunthard HF, Aberg JA, Eron JJ, et al. Antiretroviral treatment of adult HIV infection: 2014 recommendations of the International Antiviral Society‐USA Panel. JAMA. 2014;312:410–425. - PubMed
-
- World Health Organization . Global AIDS Update. Geneva: World Health Organization; 2016. Available at: www.who.int/hiv/pub/arv/global-aids-update-2016-pub/en/. Accessed June, 2016.
-
- Bates M, Mudenda V, Shibemba A, et al. Burden of tuberculosis at post mortem in inpatients at a tertiary referral centre in sub‐Saharan Africa: a prospective descriptive autopsy study. Lancet Infect Dis. 2015;15:544–551. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

