Enhanced Isolation and Release of Circulating Tumor Cells Using Nanoparticle Binding and Ligand Exchange in a Microfluidic Chip
- PMID: 28133963
- PMCID: PMC5506378
- DOI: 10.1021/jacs.6b12236
Enhanced Isolation and Release of Circulating Tumor Cells Using Nanoparticle Binding and Ligand Exchange in a Microfluidic Chip
Abstract
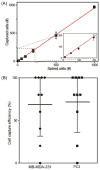
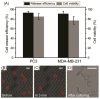
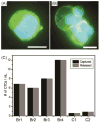
The detection of rare circulating tumor cells (CTCs) in the blood of cancer patients has the potential to be a powerful and noninvasive method for examining metastasis, evaluating prognosis, assessing tumor sensitivity to drugs, and monitoring therapeutic outcomes. In this study, we have developed an efficient strategy to isolate CTCs from the blood of breast cancer patients using a microfluidic immune-affinity approach. Additionally, to gain further access to these rare cells for downstream characterization, our strategy allows for easy detachment of the captured CTCs from the substrate without compromising cell viability or the ability to employ next generation RNA sequencing for the identification of specific breast cancer genes. To achieve this, a chemical ligand-exchange reaction was engineered to release cells attached to a gold nanoparticle coating bound to the surface of a herringbone microfluidic chip (NP-HBCTC-Chip). Compared to the use of the unmodified HBCTC-Chip, our approach provides several advantages, including enhanced capture efficiency and recovery of isolated CTCs.
Conflict of interest statement
The authors declare no competing financial interest.
Figures





Similar articles
-
Two-stage microfluidic chip for selective isolation of circulating tumor cells (CTCs).Biosens Bioelectron. 2015 May 15;67:86-92. doi: 10.1016/j.bios.2014.07.019. Epub 2014 Jul 14. Biosens Bioelectron. 2015. PMID: 25060749
-
Dual-Multivalent-Aptamer-Conjugated Nanoprobes for Superefficient Discerning of Single Circulating Tumor Cells in a Microfluidic Chip with Inductively Coupled Plasma Mass Spectrometry Detection.ACS Appl Mater Interfaces. 2021 Sep 15;13(36):43668-43675. doi: 10.1021/acsami.1c11953. Epub 2021 Sep 2. ACS Appl Mater Interfaces. 2021. PMID: 34473482
-
Isolation of circulating tumor cells using a microvortex-generating herringbone-chip.Proc Natl Acad Sci U S A. 2010 Oct 26;107(43):18392-7. doi: 10.1073/pnas.1012539107. Epub 2010 Oct 7. Proc Natl Acad Sci U S A. 2010. PMID: 20930119 Free PMC article.
-
[Advances in isolation and enrichment of circulating tumor cells in microfluidic chips].Se Pu. 2014 Jan;32(1):7-12. doi: 10.3724/sp.j.1123.2013.08009. Se Pu. 2014. PMID: 24783862 Review. Chinese.
-
Microfluidic technologies.Recent Results Cancer Res. 2012;195:59-67. doi: 10.1007/978-3-642-28160-0_5. Recent Results Cancer Res. 2012. PMID: 22527494 Review.
Cited by
-
Presence of circulating tumor cells is associated with metabolic-related variables in postoperative patients with early-stage breast cancer.Chin J Cancer Res. 2018 Jun;30(3):340-350. doi: 10.21147/j.issn.1000-9604.2018.03.06. Chin J Cancer Res. 2018. PMID: 30046228 Free PMC article.
-
An Immune-Magnetophoretic Device for the Selective and Precise Enrichment of Circulating Tumor Cells from Whole Blood.Micromachines (Basel). 2020 May 30;11(6):560. doi: 10.3390/mi11060560. Micromachines (Basel). 2020. PMID: 32486306 Free PMC article.
-
Liquid biopsies to occult brain metastasis.Mol Cancer. 2022 May 10;21(1):113. doi: 10.1186/s12943-022-01577-x. Mol Cancer. 2022. PMID: 35538484 Free PMC article. Review.
-
Capture and Selective Release of Viable Circulating Tumor Cells.Methods Mol Biol. 2023;2679:67-81. doi: 10.1007/978-1-0716-3271-0_5. Methods Mol Biol. 2023. PMID: 37300609
-
Integrated "lab-on-a-chip" microfluidic systems for isolation, enrichment, and analysis of cancer biomarkers.Lab Chip. 2023 Jun 28;23(13):2942-2958. doi: 10.1039/d2lc01076c. Lab Chip. 2023. PMID: 37314731 Free PMC article. Review.
References
-
- Neki K, Kawahara H, Watanabe K, Toyama Y, Akiba T, Yanaga K. Anticancer Res. 2013;33:1769–1772. - PubMed
- Cohen SJ, Punt CJA, Iannotti N, Saidman BH, Sabbath KD, Gabrail NY, Picus J, Morse M, Mitchell E, Miller MC, Doyle GV, Tissing H, Terstappen L, Meropol NJ. J Clin Oncol. 2008;26:3213–3221. - PubMed
- Yoon HJ, Kozminsky M, Nagrath S. ACS Nano. 2014;8:1995–2017. - PMC - PubMed
-
- Pantel K, Brakenhoff RH, Brandt B. Nat Rev Cancer. 2008;8:329–40. - PubMed
- Cristofanilli M, Budd GT, Ellis MJ, Stopeck A, Matera J, Miller MC, Reuben JM, Doyle GV, Allard WJ, Terstappen LW, Hayes DF. N Engl J Med. 2004;351:781–91. - PubMed
- Lianidou ES, Strati A, Markou A. Crit Rev Clin Lab Sci. 2014;51:160–71. - PubMed
-
- Obermayr E, Castillo-Tong DC, Pils D, Speiser P, Braicu I, Van Gorp T, Mahner S, Sehouli J, Vergote I, Zeillinger R. Gynecol Oncol. 2013;128:15–21. - PubMed
-
- Fong ZV, Winter JM. Cancer J. 2012;18:530–538. - PubMed
-
- Ashworth TR. Aust Med J. 1869;14:146–149.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

