Specialized interfaces of Smc5/6 control hinge stability and DNA association
- PMID: 28134253
- PMCID: PMC5290277
- DOI: 10.1038/ncomms14011
Specialized interfaces of Smc5/6 control hinge stability and DNA association
Abstract
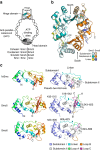
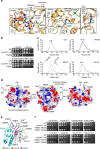
The Structural Maintenance of Chromosomes (SMC) complexes: cohesin, condensin and Smc5/6 are involved in the organization of higher-order chromosome structure-which is essential for accurate chromosome duplication and segregation. Each complex is scaffolded by a specific SMC protein dimer (heterodimer in eukaryotes) held together via their hinge domains. Here we show that the Smc5/6-hinge, like those of cohesin and condensin, also forms a toroidal structure but with distinctive subunit interfaces absent from the other SMC complexes; an unusual 'molecular latch' and a functional 'hub'. Defined mutations in these interfaces cause severe phenotypic effects with sensitivity to DNA-damaging agents in fission yeast and reduced viability in human cells. We show that the Smc5/6-hinge complex binds preferentially to ssDNA and that this interaction is affected by both 'latch' and 'hub' mutations, suggesting a key role for these unique features in controlling DNA association by the Smc5/6 complex.
Figures





References
-
- Ju L. et al. SMC6 is an essential gene in mice, but a hypomorphic mutant in the ATPase domain has a mild phenotype with a range of subtle abnormalities. DNA Repair (Amst) 12, 356–366 (2013). - PubMed
-
- Branzei D. et al. Ubc9- and mms21-mediated sumoylation counteracts recombinogenic events at damaged replication forks. Cell 127, 509–522 (2006). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

