Transcriptomic and anatomic parcellation of 5-HT3AR expressing cortical interneuron subtypes revealed by single-cell RNA sequencing
- PMID: 28134272
- PMCID: PMC5290279
- DOI: 10.1038/ncomms14219
Transcriptomic and anatomic parcellation of 5-HT3AR expressing cortical interneuron subtypes revealed by single-cell RNA sequencing
Abstract
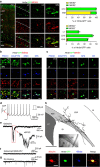
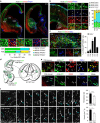
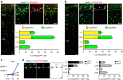
Cortical GABAergic interneurons constitute a highly diverse population of inhibitory neurons that are key regulators of cortical microcircuit function. An important and heterogeneous group of cortical interneurons specifically expresses the serotonin receptor 3A (5-HT3AR) but how this diversity emerges during development is poorly understood. Here we use single-cell transcriptomics to identify gene expression patterns operating in Htr3a-GFP+ interneurons during early steps of cortical circuit assembly. We identify three main molecular types of Htr3a-GFP+ interneurons, each displaying distinct developmental dynamics of gene expression. The transcription factor Meis2 is specifically enriched in a type of Htr3a-GFP+ interneurons largely confined to the cortical white matter. These MEIS2-expressing interneurons appear to originate from a restricted region located at the embryonic pallial-subpallial boundary. Overall, this study identifies MEIS2 as a subclass-specific marker for 5-HT3AR-containing interstitial interneurons and demonstrates that the transcriptional and anatomical parcellation of cortical interneurons is developmentally coupled.
Conflict of interest statement
The authors declare no competing financial interests.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

