Computational predictors fail to identify amino acid substitution effects at rheostat positions
- PMID: 28134345
- PMCID: PMC5278360
- DOI: 10.1038/srep41329
Computational predictors fail to identify amino acid substitution effects at rheostat positions
Abstract
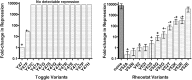
Many computational approaches exist for predicting the effects of amino acid substitutions. Here, we considered whether the protein sequence position class - rheostat or toggle - affects these predictions. The classes are defined as follows: experimentally evaluated effects of amino acid substitutions at toggle positions are binary, while rheostat positions show progressive changes. For substitutions in the LacI protein, all evaluated methods failed two key expectations: toggle neutrals were incorrectly predicted as more non-neutral than rheostat non-neutrals, while toggle and rheostat neutrals were incorrectly predicted to be different. However, toggle non-neutrals were distinct from rheostat neutrals. Since many toggle positions are conserved, and most rheostats are not, predictors appear to annotate position conservation better than mutational effect. This finding can explain the well-known observation that predictors assign disproportionate weight to conservation, as well as the field's inability to improve predictor performance. Thus, building reliable predictors requires distinguishing between rheostat and toggle positions.
Conflict of interest statement
The authors declare no competing financial interests.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

