Mutations in tropomyosin 4 underlie a rare form of human macrothrombocytopenia
- PMID: 28134622
- PMCID: PMC5330761
- DOI: 10.1172/JCI86154
Mutations in tropomyosin 4 underlie a rare form of human macrothrombocytopenia
Abstract
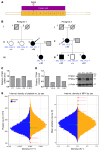
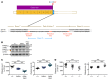
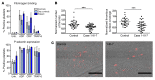
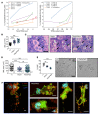
Platelets are anuclear cells that are essential for blood clotting. They are produced by large polyploid precursor cells called megakaryocytes. Previous genome-wide association studies in nearly 70,000 individuals indicated that single nucleotide variants (SNVs) in the gene encoding the actin cytoskeletal regulator tropomyosin 4 (TPM4) exert an effect on the count and volume of platelets. Platelet number and volume are independent risk factors for heart attack and stroke. Here, we have identified 2 unrelated families in the BRIDGE Bleeding and Platelet Disorders (BPD) collection who carry a TPM4 variant that causes truncation of the TPM4 protein and segregates with macrothrombocytopenia, a disorder characterized by low platelet count. N-Ethyl-N-nitrosourea-induced (ENU-induced) missense mutations in Tpm4 or targeted inactivation of the Tpm4 locus led to gene dosage-dependent macrothrombocytopenia in mice. All other blood cell counts in Tpm4-deficient mice were normal. Insufficient TPM4 expression in human and mouse megakaryocytes resulted in a defect in the terminal stages of platelet production and had a mild effect on platelet function. Together, our findings demonstrate a nonredundant role for TPM4 in platelet biogenesis in humans and mice and reveal that truncating variants in TPM4 cause a previously undescribed dominant Mendelian platelet disorder.
Conflict of interest statement
Figures




References
-
- Nieswandt B, Pleines I, Bender M. Platelet adhesion and activation mechanisms in arterial thrombosis and ischaemic stroke. J Thromb Haemost. 2011;9(suppl 1):92–104. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

