Systematic analysis of telomere length and somatic alterations in 31 cancer types
- PMID: 28135248
- PMCID: PMC5571729
- DOI: 10.1038/ng.3781
Systematic analysis of telomere length and somatic alterations in 31 cancer types
Abstract
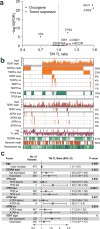
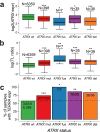
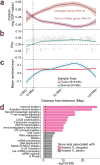
Cancer cells survive cellular crisis through telomere maintenance mechanisms. We report telomere lengths in 18,430 samples, including tumors and non-neoplastic samples, across 31 cancer types. Telomeres were shorter in tumors than in normal tissues and longer in sarcomas and gliomas than in other cancers. Among 6,835 cancers, 73% expressed telomerase reverse transcriptase (TERT), which was associated with TERT point mutations, rearrangements, DNA amplifications and transcript fusions and predictive of telomerase activity. TERT promoter methylation provided an additional deregulatory TERT expression mechanism. Five percent of cases, characterized by undetectable TERT expression and alterations in ATRX or DAXX, demonstrated elongated telomeres and increased telomeric repeat-containing RNA (TERRA). The remaining 22% of tumors neither expressed TERT nor harbored alterations in ATRX or DAXX. In this group, telomere length positively correlated with TP53 and RB1 mutations. Our analysis integrates TERT abnormalities, telomerase activity and genomic alterations with telomere length in cancer.
Conflict of interest statement
The authors declare no competing financial interests.
Figures



References
-
- Olovnikov AM. A theory of marginotomy. The incomplete copying of template margin in enzymic synthesis of polynucleotides and biological significance of the phenomenon. J Theor Biol. 1973;41:181–90. - PubMed
-
- Shay JW, Pereira-Smith OM, Wright WE. A role for both RB and p53 in the regulation of human cellular senescence. Exp Cell Res. 1991;196:33–9. - PubMed
-
- Stewart SA, Weinberg RA. Telomeres: cancer to human aging. Annu Rev Cell Dev Biol. 2006;22:531–57. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

