Massively multiplex single-cell Hi-C
- PMID: 28135255
- PMCID: PMC5330809
- DOI: 10.1038/nmeth.4155
Massively multiplex single-cell Hi-C
Abstract
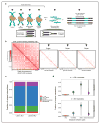
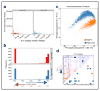
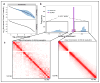
We present single-cell combinatorial indexed Hi-C (sciHi-C), a method that applies combinatorial cellular indexing to chromosome conformation capture. In this proof of concept, we generate and sequence six sciHi-C libraries comprising a total of 10,696 single cells. We use sciHi-C data to separate cells by karyotypic and cell-cycle state differences and identify cell-to-cell heterogeneity in mammalian chromosomal conformation. Our results demonstrate that combinatorial indexing is a generalizable strategy for single-cell genomics.
Conflict of interest statement
K.G. and F.S. are employees of Illumina Inc.
Figures
Comment in
-
Technique: Barcoding the nucleus.Nat Rev Genet. 2017 Apr;18(4):211. doi: 10.1038/nrg.2017.11. Epub 2017 Feb 13. Nat Rev Genet. 2017. PMID: 28190875 No abstract available.
-
Stratifying tissue heterogeneity with scalable single-cell assays.Nat Methods. 2017 Feb 28;14(3):238-239. doi: 10.1038/nmeth.4209. Nat Methods. 2017. PMID: 28245217 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

