Hypoxia Sensing and Persistence Genes Are Expressed during the Intragranulomatous Survival of Mycobacterium tuberculosis
- PMID: 28135421
- PMCID: PMC5449490
- DOI: 10.1165/rcmb.2016-0239OC
Hypoxia Sensing and Persistence Genes Are Expressed during the Intragranulomatous Survival of Mycobacterium tuberculosis
Abstract
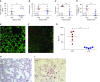
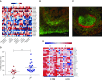
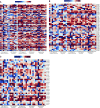
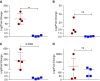
Although it is accepted that the environment within the granuloma profoundly affects Mycobacterium tuberculosis (Mtb) and infection outcome, our ability to understand Mtb gene expression in these niches has been limited. We determined intragranulomatous gene expression in human-like lung lesions derived from nonhuman primates with both active tuberculosis (ATB) and latent TB infection (LTBI). We employed a non-laser-based approach to microdissect individual lung lesions and interrogate the global transcriptome of Mtb within granulomas. Mtb genes expressed in classical granulomas with central, caseous necrosis, as well as within the caseum itself, were identified and compared with other Mtb lesions in animals with ATB (n = 7) or LTBI (n = 7). Results were validated using both an oligonucleotide approach and RT-PCR on macaque samples and by using human TB samples. We detected approximately 2,900 and 1,850 statistically significant genes in ATB and LTBI lesions, respectively (linear models for microarray analysis, Bonferroni corrected, P < 0.05). Of these genes, the expression of approximately 1,300 (ATB) and 900 (LTBI) was positively induced. We identified the induction of key regulons and compared our results to genes previously determined to be required for Mtb growth. Our results indicate pathways that Mtb uses to ensure its survival in a highly stressful environment in vivo. A large number of genes is commonly expressed in granulomas with ATB and LTBI. In addition, the enhanced expression of the dormancy survival regulon was a key feature of lesions in animals with LTBI, stressing its importance in the persistence of Mtb during the chronic phase of infection.
Keywords: Mycobacterium tuberculosis; granuloma; hypoxia; macaque; microdissection.
Figures


References
-
- Foreman TW, Mehra S, LoBato DN, Malek A, Alvarez X, Golden NA, Bucşan AN, Didier PJ, Doyle-Meyers LA, Russell-Lodrigue KE, et al. CD4+ T-cell-independent mechanisms suppress reactivation of latent tuberculosis in a macaque model of HIV coinfection. Proc Natl Acad Sci USA. 2016;113(38):E5636–E5644. - PMC - PubMed
-
- Mehra S, Golden NA, Stuckey K, Didier PJ, Doyle LA, Russell-Lodrigue KE, Sugimoto C, Hasegawa A, Sivasubramani SK, Roy CJ, et al. The Mycobacterium tuberculosis stress response factor SigH is required for bacterial burden as well as immunopathology in primate lungs. J Infect Dis. 2012;205:1203–1213. - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

