CRISPR-Cas type II-based Synthetic Biology applications in eukaryotic cells
- PMID: 28136159
- PMCID: PMC5711462
- DOI: 10.1080/15476286.2017.1282024
CRISPR-Cas type II-based Synthetic Biology applications in eukaryotic cells
Abstract
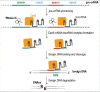
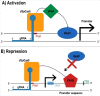
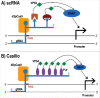
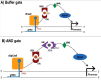
The CRISPR-Cas system has rapidly reached a huge popularity as a new, powerful method for precise DNA editing and genome reengineering. In Synthetic Biology, the CRISPR-Cas type II system has inspired the construction of a novel class of RNA-based transcription factors. In their simplest form, they are made of a CRISPR RNA molecule, which targets a promoter sequence, and a deficient Cas9 (i.e. deprived of any nuclease activity) that has been fused to an activation or a repression domain. Up- and downregulation of single genes in mammalian and yeast cells have been achieved with satisfactory results. Moreover, the construction of CRISPR-based transcription factors is much simpler than the assembly of synthetic proteins such as the Transcription Activator-Like effectors. However, the feasibility of complex synthetic networks fully based on the CRISPR-dCas9 technology has still to be proved and new designs, which take into account different CRISPR types, shall be investigated.
Keywords: CRISPR; Cas9; Synthetic Biology; gene circuits; guide RNA.
Figures

References
-
- Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science 2007; 315(5819):1709-12; PMID:17379808; https://doi.org/ 10.1126/science.1138140 - DOI - PubMed
-
- Horvath P, Barrangou R. CRISPR/Cas, the immune system of bacteria and archaea. Science 2010; 327(5962):167-70; PMID:20056882; https://doi.org/ 10.1126/science.1179555 - DOI - PubMed
-
- Marraffini LA, Sontheimer EJ. CRISPR interference: RNA-directed adaptive immunity in bacteria and archaea. Nat Rev Genet 2010; 11(3):181-190; PMID:20125085; https://doi.org/ 10.1038/nrg2749 - DOI - PMC - PubMed
-
- Wright AV, Nuñez JK, Doudna JA. Biology and applications of CRISPR systems: harnessing nature's toolbox for genome engineering. Cell 2016; 164(1–2):29-44; PMID:26771484; https://doi.org/ 10.1016/j.cell.2015.12.035 - DOI - PubMed
-
- Garneau JE, Dupuis MÈ, Villion M, Romero DA, Barrangou R, Boyaval P, Fremaux C, Horvath P, Magadán AH, Moineau S. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature 2010; 468(7320):67-71; PMID:21048762; https://doi.org/ 10.1038/nature09523 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
