Epithelial response to a high-protein diet in rat colon
- PMID: 28137254
- PMCID: PMC5282643
- DOI: 10.1186/s12864-017-3514-z
Epithelial response to a high-protein diet in rat colon
Abstract
Background: High-protein diets (HPD) alter the large intestine microbiota composition in association with a metabolic shift towards protein degradation. Some amino acid-derived metabolites produced by the colon bacteria are beneficial for the mucosa while others are deleterious at high concentrations. The aim of the present work was to define the colonic epithelial response to an HPD. Transcriptome profiling was performed on colonocytes of rats fed an HPD or an isocaloric normal-protein diet (NPD) for 2 weeks.
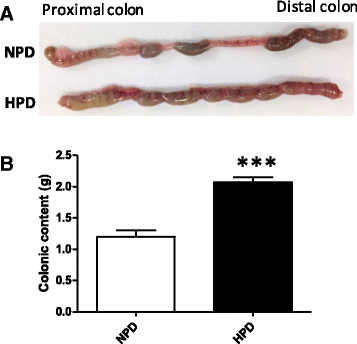
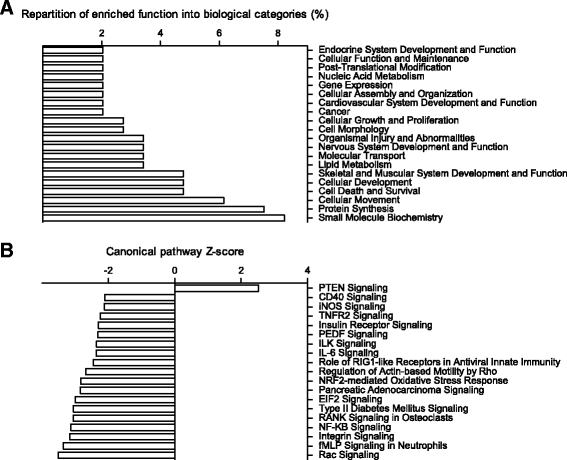
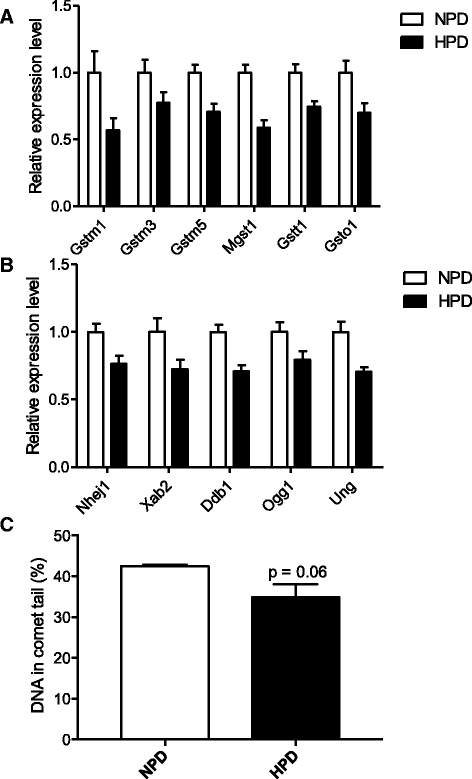
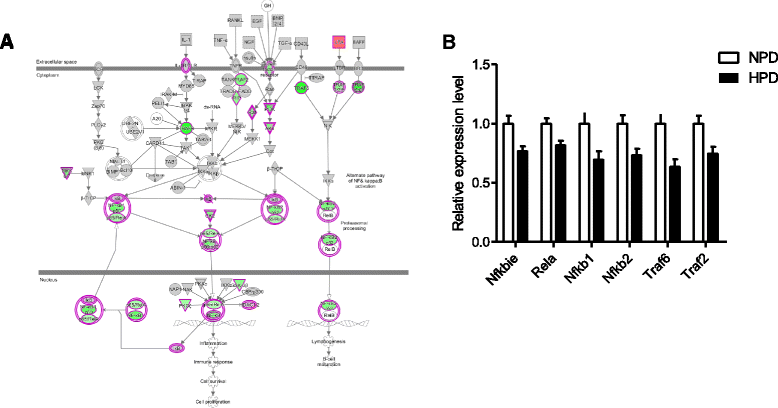
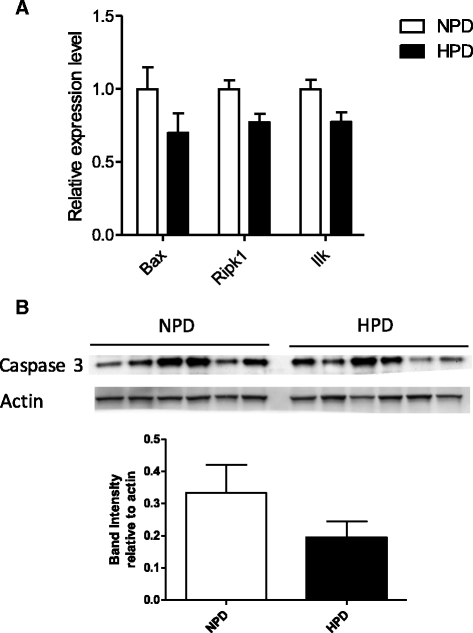
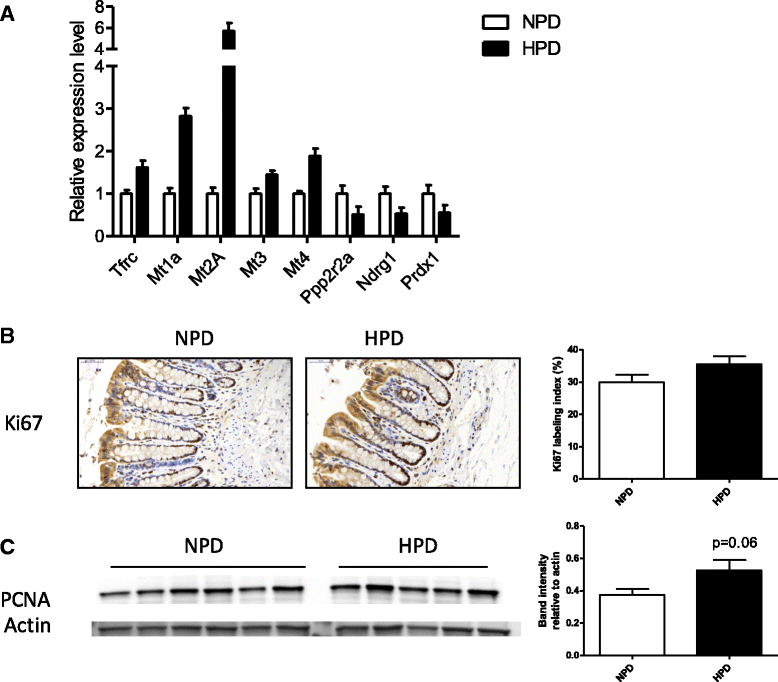
Results: The HPD downregulated the expression of genes notably implicated in pathways related to cellular metabolism, NF-κB signaling, DNA repair, glutathione metabolism and cellular adhesion in colonocytes. In contrast, the HPD upregulated the expression of genes related to cell proliferation and chemical barrier function. These changes at the mRNA level in colonocytes were not associated with detrimental effects of the HPD on DNA integrity (comet assay), epithelium renewal (quantification of proliferation and apoptosis markers by immunohistochemistry and western blot) and colonic barrier integrity (Ussing chamber experiments).
Conclusion: The modifications of the luminal environment after an HPD were associated with maintenance of the colonic homeostasis that might be the result of adaptive processes in the epithelium related to the observed transcriptional regulations.
Keywords: Barrier function; Colon; DNA damages; Dietary protein; Epithelial cells; Epithelial renewal; High-protein diet; Mucus; Transcriptome.
Figures

References
-
- Bax M-L, Buffière C, Hafnaoui N, Gaudichon C, Savary-Auzeloux I, Dardevet D, et al. Effects of meat cooking, and of ingested amount, on protein digestion speed and entry of residual proteins into the colon: a study in minipigs. PLoS One. 2013;8:e61252. doi: 10.1371/journal.pone.0061252. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

