Sparse network modeling and metscape-based visualization methods for the analysis of large-scale metabolomics data
- PMID: 28137712
- PMCID: PMC5860222
- DOI: 10.1093/bioinformatics/btx012
Sparse network modeling and metscape-based visualization methods for the analysis of large-scale metabolomics data
Abstract
Motivation: Recent technological advances in mass spectrometry, development of richer mass spectral libraries and data processing tools have enabled large scale metabolic profiling. Biological interpretation of metabolomics studies heavily relies on knowledge-based tools that contain information about metabolic pathways. Incomplete coverage of different areas of metabolism and lack of information about non-canonical connections between metabolites limits the scope of applications of such tools. Furthermore, the presence of a large number of unknown features, which cannot be readily identified, but nonetheless can represent bona fide compounds, also considerably complicates biological interpretation of the data.
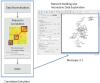
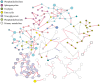
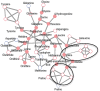
Results: Leveraging recent developments in the statistical analysis of high-dimensional data, we developed a new Debiased Sparse Partial Correlation algorithm (DSPC) for estimating partial correlation networks and implemented it as a Java-based CorrelationCalculator program. We also introduce a new version of our previously developed tool Metscape that enables building and visualization of correlation networks. We demonstrate the utility of these tools by constructing biologically relevant networks and in aiding identification of unknown compounds.
Availability and implementation: http://metscape.med.umich.edu.
Supplementary information: Supplementary data are available at Bioinformatics online.
© The Author 2017. Published by Oxford University Press. All rights reserved. For Permissions, please e-mail: journals.permissions@oup.com
Figures



References
-
- Alonso A. et al. (2011) AStream: an R package for annotating LC/MS metabolomic data. Bioinformatics, 27, 1339–1340. - PubMed
-
- Benjamini Y., Hochberg Y. (1995) Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. J. Roy. Stat. Soc. B Met., 57, 289–300.
-
- Bühlmann P., Van De Geer S.. Statistics for High-Dimensional Data: Methods, Theory and Applications. Springer Science & Business Media, 2011.
MeSH terms
Grants and funding
- U01 AG012554/AG/NIA NIH HHS/United States
- R21 GM101719/GM/NIGMS NIH HHS/United States
- P30 DK089503/DK/NIDDK NIH HHS/United States
- U01 AG012535/AG/NIA NIH HHS/United States
- U01 AG012553/AG/NIA NIH HHS/United States
- U01 NR004061/NR/NINR NIH HHS/United States
- U01 AG012539/AG/NIA NIH HHS/United States
- U01 AG012546/AG/NIA NIH HHS/United States
- U01 AG012495/AG/NIA NIH HHS/United States
- U01 AG012505/AG/NIA NIH HHS/United States
- R03 CA211817/CA/NCI NIH HHS/United States
- U01 AG012531/AG/NIA NIH HHS/United States
- U01 AG017719/AG/NIA NIH HHS/United States
- R01 GM114029/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources

