Arabidopsis HAP2/GCS1 is a gamete fusion protein homologous to somatic and viral fusogens
- PMID: 28137780
- PMCID: PMC5350521
- DOI: 10.1083/jcb.201610093
Arabidopsis HAP2/GCS1 is a gamete fusion protein homologous to somatic and viral fusogens
Abstract
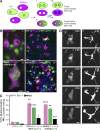
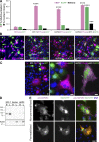
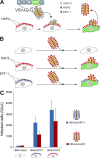
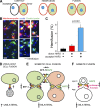
Cell-cell fusion is inherent to sexual reproduction. Loss of HAPLESS 2/GENERATIVE CELL SPECIFIC 1 (HAP2/GCS1) proteins results in gamete fusion failure in diverse organisms, but their exact role is unclear. In this study, we show that Arabidopsis thaliana HAP2/GCS1 is sufficient to promote mammalian cell-cell fusion. Hemifusion and complete fusion depend on HAP2/GCS1 presence in both fusing cells. Furthermore, expression of HAP2 on the surface of pseudotyped vesicular stomatitis virus results in homotypic virus-cell fusion. We demonstrate that the Caenorhabditis elegans Epithelial Fusion Failure 1 (EFF-1) somatic cell fusogen can replace HAP2/GCS1 in one of the fusing membranes, indicating that HAP2/GCS1 and EFF-1 share a similar fusion mechanism. Structural modeling of the HAP2/GCS1 protein family predicts that they are homologous to EFF-1 and viral class II fusion proteins (e.g., Zika virus). We name this superfamily Fusexins: fusion proteins essential for sexual reproduction and exoplasmic merger of plasma membranes. We suggest a common origin and evolution of sexual reproduction, enveloped virus entry into cells, and somatic cell fusion.
© 2017 Valansi et al.
Figures

Comment on
-
What Came First-the Virus or the Egg?Cell. 2017 Feb 23;168(5):755-757. doi: 10.1016/j.cell.2017.02.012. Cell. 2017. PMID: 28235193
References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

