A Whole-Genome Sequencing Approach To Study Cefoxitin-Resistant Salmonella enterica Serovar Heidelberg Isolates from Various Sources
- PMID: 28137797
- PMCID: PMC5365727
- DOI: 10.1128/AAC.01919-16
A Whole-Genome Sequencing Approach To Study Cefoxitin-Resistant Salmonella enterica Serovar Heidelberg Isolates from Various Sources
Abstract
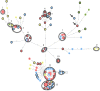
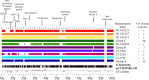
This study characterized cefoxitin-resistant and -susceptible Salmonella enterica serovar Heidelberg strains from humans, abattoir poultry, and retail poultry to assess the molecular relationships of isolates from these sources in Québec in 2012. Isolates were collected as part of the Canadian Integrated Program for Antimicrobial Resistance Surveillance (CIPARS). All isolates were subjected to antimicrobial susceptibility testing, PCR for CMY-2, pulsed-field gel electrophoresis (PFGE), and whole-genome sequencing (WGS). A total of 113 S Heidelberg isolates from humans (n = 51), abattoir poultry (n = 18), and retail poultry (n = 44) were studied. All cefoxitin-resistant isolates (n = 65) were also resistant to amoxicillin-clavulanic acid, ampicillin, ceftiofur, and ceftriaxone, and all contained the CMY-2 gene. PFGE analysis showed that 111/113 (98.2%) isolates clustered together with ≥90% similarity. Core genome analysis using WGS identified 13 small clusters of isolates with 0 to 4 single nucleotide variations (SNVs), consisting of cefoxitin-resistant and -susceptible human, abattoir poultry, and retail poultry isolates. CMY-2 plasmids from cefoxitin-resistant isolates all belonged to incompatibility group I1. Analysis of IncI1 plasmid sequences revealed high identity (95 to 99%) to a previously described plasmid (pCVM29188_101) found in Salmonella Kentucky. When compared to pCVM29188_101, all sequenced cefoxitin-resistant isolates were found to carry 1 of 10 possible variant plasmids. Transmission of S Heidelberg may be occurring between human, abattoir poultry, and retail poultry sources, and transmission of a common CMY-2 plasmid may be occurring among S Heidelberg strains with variable genetic backgrounds.
Keywords: Québec; Salmonella Heidelberg; beta-lactamase; cefoxitin; plasmid analysis; whole-genome sequencing.
© Crown copyright 2017.
Figures

References
-
- Public Health Agency of Canada. 2014. National Enteric Surveillance Program (NESP) annual report 2014. http://publications.gc.ca/collections/collection_2016/aspc-phac/HP37-15-....
-
- Public Health Agency of Canada. 2012. National Enteric Surveillance Program (NESP) annual report 2012. http://publications.gc.ca/collections/collection_2014/aspc-phac/HP37-15-....
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

