Binding, sliding, and function of cohesin during transcriptional activation
- PMID: 28137853
- PMCID: PMC5320966
- DOI: 10.1073/pnas.1617309114
Binding, sliding, and function of cohesin during transcriptional activation
Abstract
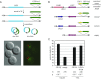
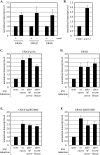
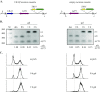
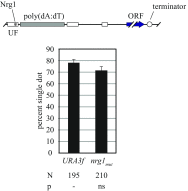
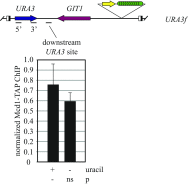
The ring-shaped cohesin complex orchestrates long-range DNA interactions to mediate sister chromatid cohesion and other aspects of chromosome structure and function. In the yeast Saccharomyces cerevisiae, the complex binds discrete sites along chromosomes, including positions within and around genes. Transcriptional activity redistributes the complex to the 3' ends of convergently oriented gene pairs. Despite the wealth of information about where cohesin binds, little is known about cohesion at individual chromosomal binding sites and how transcription affects cohesion when cohesin complexes redistribute. In this study, we generated extrachromosomal DNA circles to study cohesion in response to transcriptional induction of a model gene, URA3. Functional cohesin complexes loaded onto the locus via a poly(dA:dT) tract in the gene promoter and mediated cohesion before induction. Upon transcription, the fate of these complexes depended on whether the DNA was circular or not. When gene activation occurred before DNA circularization, cohesion was lost. When activation occurred after DNA circularization, cohesion persisted. The presence of a convergently oriented gene also prevented transcription-driven loss of functional cohesin complexes, at least in M phase-arrested cells. The results are consistent with cohesin binding chromatin in a topological embrace and with transcription mobilizing functional complexes by sliding them along DNA.
Keywords: URA3; cohesin; poly(dA:dT); sister chromatid cohesion; transcription.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






References
-
- Michaelis C, Ciosk R, Nasmyth K. Cohesins: Chromosomal proteins that prevent premature separation of sister chromatids. Cell. 1997;91(1):35–45. - PubMed
-
- Cucco F, Musio A. Genome stability: What we have learned from cohesinopathies. Am J Med Genet C Semin Med Genet. 2016;172(2):171–178. - PubMed
-
- Onn I, Heidinger-Pauli JM, Guacci V, Ünal E, Koshland DE. Sister chromatid cohesion: A simple concept with a complex reality. Annu Rev Cell Dev Biol. 2008;24:105–129. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

