Effects of Mutations on the Aggregation Propensity of the Human Prion-Like Protein hnRNPA2B1
- PMID: 28137911
- PMCID: PMC5376634
- DOI: 10.1128/MCB.00652-16
Effects of Mutations on the Aggregation Propensity of the Human Prion-Like Protein hnRNPA2B1
Abstract
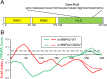
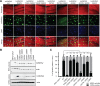
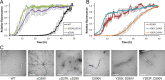
Hundreds of human proteins contain prion-like domains, which are a subset of low-complexity domains with high amino acid compositional similarity to yeast prion domains. A recently characterized mutation in the prion-like domain of the human heterogeneous nuclear ribonucleoprotein hnRNPA2B1 increases the aggregation propensity of the protein and causes multisystem proteinopathy. The mutant protein forms cytoplasmic inclusions when expressed in Drosophila, the mutation accelerates aggregation in vitro, and the mutant prion-like domain can substitute for a portion of a yeast prion domain in supporting prion activity. To examine the relationship between amino acid sequence and aggregation propensity, we made a diverse set of point mutations in the hnRNPA2B1 prion-like domain. We found that the effects on prion formation in Saccharomyces cerevisiae and aggregation in vitro could be predicted entirely based on amino acid composition. However, composition was an imperfect predictor of inclusion formation in Drosophila; while most mutations showed similar behaviors in yeast, in vitro, and in Drosophila, a few showed anomalous behavior. Collectively, these results demonstrate the significant progress that has been made in predicting the effects of mutations on intrinsic aggregation propensity while also highlighting the challenges of predicting the effects of mutations in more complex organisms.
Keywords: aggregation; low-complexity domain; prion-like; prions.
Copyright © 2017 American Society for Microbiology.
Figures




Similar articles
-
Aggregation and degradation scales for prion-like domains: sequence features and context weigh in.Curr Genet. 2019 Apr;65(2):387-392. doi: 10.1007/s00294-018-0890-0. Epub 2018 Oct 11. Curr Genet. 2019. PMID: 30310993 Review.
-
Manipulating the aggregation activity of human prion-like proteins.Prion. 2017 Sep 3;11(5):323-331. doi: 10.1080/19336896.2017.1356560. Prion. 2017. PMID: 28934062 Free PMC article.
-
Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS.Nature. 2013 Mar 28;495(7442):467-73. doi: 10.1038/nature11922. Epub 2013 Mar 3. Nature. 2013. PMID: 23455423 Free PMC article.
-
Perfecting prediction of mutational impact on the aggregation propensity of the ALS-associated hnRNPA2 prion-like protein.FEBS Lett. 2017 Jul;591(13):1966-1971. doi: 10.1002/1873-3468.12698. Epub 2017 Jun 18. FEBS Lett. 2017. PMID: 28542905
-
Predicting the aggregation propensity of prion sequences.Virus Res. 2015 Sep 2;207:127-35. doi: 10.1016/j.virusres.2015.03.001. Epub 2015 Mar 6. Virus Res. 2015. PMID: 25747492 Review.
Cited by
-
Extremely Amyloidogenic Single-Chain Analogues of Insulin's H-Fragment: Structural Adaptability of an Amyloid Stretch.Langmuir. 2020 Oct 20;36(41):12150-12159. doi: 10.1021/acs.langmuir.0c01747. Epub 2020 Oct 7. Langmuir. 2020. PMID: 32988199 Free PMC article.
-
Analyzing the Effects of Single Nucleotide Polymorphisms on hnRNPA2/B1 Protein Stability and Function: Insights for Anticancer Therapeutic Design.ACS Omega. 2024 Jan 26;9(5):5485-5495. doi: 10.1021/acsomega.3c07195. eCollection 2024 Feb 6. ACS Omega. 2024. PMID: 38343990 Free PMC article.
-
Functional Mammalian Amyloids and Amyloid-Like Proteins.Life (Basel). 2020 Aug 21;10(9):156. doi: 10.3390/life10090156. Life (Basel). 2020. PMID: 32825636 Free PMC article. Review.
-
Aggregation and degradation scales for prion-like domains: sequence features and context weigh in.Curr Genet. 2019 Apr;65(2):387-392. doi: 10.1007/s00294-018-0890-0. Epub 2018 Oct 11. Curr Genet. 2019. PMID: 30310993 Review.
-
Defining the role of the polyasparagine repeat domain of the S. cerevisiae transcription factor Azf1p.PLoS One. 2021 May 21;16(5):e0247285. doi: 10.1371/journal.pone.0247285. eCollection 2021. PLoS One. 2021. PMID: 34019539 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
