Ribosome dynamics during decoding
- PMID: 28138068
- PMCID: PMC5311926
- DOI: 10.1098/rstb.2016.0182
Ribosome dynamics during decoding
Abstract
Elongation factors Tu (EF-Tu) and SelB are translational GTPases that deliver aminoacyl-tRNAs (aa-tRNAs) to the ribosome. In each canonical round of translation elongation, aa-tRNAs, assisted by EF-Tu, decode mRNA codons and insert the respective amino acid into the growing peptide chain. Stop codons usually lead to translation termination; however, in special cases UGA codons are recoded to selenocysteine (Sec) with the help of SelB. Recruitment of EF-Tu and SelB together with their respective aa-tRNAs to the ribosome is a multistep process. In this review, we summarize recent progress in understanding the role of ribosome dynamics in aa-tRNA selection. We describe the path to correct codon recognition by canonical elongator aa-tRNA and Sec-tRNASec and discuss the local and global rearrangements of the ribosome in response to correct and incorrect aa-tRNAs. We present the mechanisms of GTPase activation and GTP hydrolysis of EF-Tu and SelB and summarize what is known about the accommodation of aa-tRNA on the ribosome after its release from the elongation factor. We show how ribosome dynamics ensures high selectivity for the cognate aa-tRNA and suggest that conformational fluctuations, induced fit and kinetic discrimination play major roles in maintaining the speed and fidelity of translation.This article is part of the themed issue 'Perspectives on the ribosome'.
Keywords: decoding; recoding; ribosome; tRNA; translation.
© 2017 The Authors.
Figures
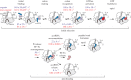




 correspond to structural intermediates resolved by cryo-EM; state
correspond to structural intermediates resolved by cryo-EM; state  was modelled based on the structural data [13].
was modelled based on the structural data [13].  The initial complex contains an mRNA with a UGA stop codon in the A site and a SECIS element exposed for SelB recruitment. The ribosome is in the classical state with a peptidyl-tRNA in the P site and a vacant A site, while the universally conserved bases A1492 and A1493 of 16S rRNA fluctuate between the flipped-in and flipped-out conformations.
The initial complex contains an mRNA with a UGA stop codon in the A site and a SECIS element exposed for SelB recruitment. The ribosome is in the classical state with a peptidyl-tRNA in the P site and a vacant A site, while the universally conserved bases A1492 and A1493 of 16S rRNA fluctuate between the flipped-in and flipped-out conformations.  The recruitment state is formed upon binding of SelB–GTP–Sec-tRNASec to the SECIS. The contact between SelB domain 4 and the SECIS is maintained in all subsequent steps. Sec-tRNASec can spontaneously sample a broad range of different conformations.
The recruitment state is formed upon binding of SelB–GTP–Sec-tRNASec to the SECIS. The contact between SelB domain 4 and the SECIS is maintained in all subsequent steps. Sec-tRNASec can spontaneously sample a broad range of different conformations.  In the transient initial binding state, Sec-tRNASec binds to the SRL and SelB to the shoulder domain of the 30S subunit. The shoulder domain moves apart stabilizing A1492 and A1493 in a flipped-in conformation.
In the transient initial binding state, Sec-tRNASec binds to the SRL and SelB to the shoulder domain of the 30S subunit. The shoulder domain moves apart stabilizing A1492 and A1493 in a flipped-in conformation.  The distance to the UGA codon decreases as tRNASec attempts to read the codon in the transient codon reading state.
The distance to the UGA codon decreases as tRNASec attempts to read the codon in the transient codon reading state.  GTPase-activated pre-hydrolysis state. Codon recognition by tRNASec induces a local closure of the decoding centre with A1492 and A1493 flipping out. The resulting global closure of the shoulder domain leads to repositioning of the tRNA and docking of SelB on the SRL. The docking leads to a codon-dependent GTPase activation of SelB (dark red shading) and GTP hydrolysis. Notably, SelB, which initially interacts with the 30S shoulder only, does not interact with the SRL until the final GTPase-activated state is reached.
GTPase-activated pre-hydrolysis state. Codon recognition by tRNASec induces a local closure of the decoding centre with A1492 and A1493 flipping out. The resulting global closure of the shoulder domain leads to repositioning of the tRNA and docking of SelB on the SRL. The docking leads to a codon-dependent GTPase activation of SelB (dark red shading) and GTP hydrolysis. Notably, SelB, which initially interacts with the 30S shoulder only, does not interact with the SRL until the final GTPase-activated state is reached.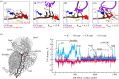

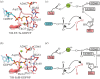
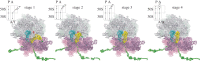
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

