Chromosome-Range Whole-Genome High-Throughput Experimental Haplotyping by Single-Chromosome Microdissection
- PMID: 28138846
- PMCID: PMC6372095
- DOI: 10.1007/978-1-4939-6750-6_9
Chromosome-Range Whole-Genome High-Throughput Experimental Haplotyping by Single-Chromosome Microdissection
Abstract
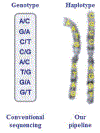
Haplotype is fundamental genetic information; it provides essential information for deciphering the functional and etiological roles of genetic variants. As haplotype information is closely related to the functional and etiological impact of genetic variants, it is widely anticipated that haplotype information will be extremely valuable in a wide spectra of applications, including academic research, clinical diagnosis of genetic disease and in the pharmaceutical industry. Haplotyping is essential for LD (linkage disequilibrium) mapping, functional studies on cis-interactions, big data imputation, association studies, population studies, and evolutionary studies. Unfortunately, current sequencing technologies and genotyping arrays do not routinely deliver this information for each individual, but yield only unphased genotypes. Here, we describe a high-throughput and cost-effective experimental protocol to obtain high-resolution chromosomal haplotypes of each individual diploid (including human) genome by the single-chromosome microdissection and sequencing approach.
Keywords: Chromosome-length; Experimental; Haplotype; High-throughput; Whole-genome.
Figures

References
-
- Liu N, Zhang K, Zhao H (2008) Haplotype- association analysis. Adv Genet 60:335–405 - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

