The PRC2-binding long non-coding RNAs in human and mouse genomes are associated with predictive sequence features
- PMID: 28139710
- PMCID: PMC5282597
- DOI: 10.1038/srep41669
The PRC2-binding long non-coding RNAs in human and mouse genomes are associated with predictive sequence features
Abstract
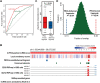
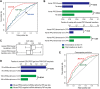
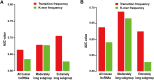
Recently, long non-coding RNAs (lncRNAs) have emerged as an important class of molecules involved in many cellular processes. One of their primary functions is to shape epigenetic landscape through interactions with chromatin modifying proteins. However, mechanisms contributing to the specificity of such interactions remain poorly understood. Here we took the human and mouse lncRNAs that were experimentally determined to have physical interactions with Polycomb repressive complex 2 (PRC2), and systematically investigated the sequence features of these lncRNAs by developing a new computational pipeline for sequences composition analysis, in which each sequence is considered as a series of transitions between adjacent nucleotides. Through that, PRC2-binding lncRNAs were found to be associated with a set of distinctive and evolutionarily conserved sequence features, which can be utilized to distinguish them from the others with considerable accuracy. We further identified fragments of PRC2-binding lncRNAs that are enriched with these sequence features, and found they show strong PRC2-binding signals and are more highly conserved across species than the other parts, implying their functional importance.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

