Functional amyloids in Streptococcus mutans, their use as targets of biofilm inhibition and initial characterization of SMU_63c
- PMID: 28141493
- PMCID: PMC5775903
- DOI: 10.1099/mic.0.000443
Functional amyloids in Streptococcus mutans, their use as targets of biofilm inhibition and initial characterization of SMU_63c
Abstract
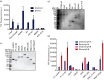
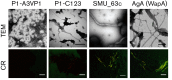
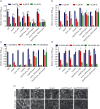
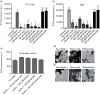
Amyloids have been identified as functional components of the extracellular matrix of bacterial biofilms. Streptococcus mutans is an established aetiologic agent of dental caries and a biofilm dweller. In addition to the previously identified amyloidogenic adhesin P1 (also known as AgI/II, PAc), we show that the naturally occurring antigen A derivative of S. mutans wall-associated protein A (WapA) and the secreted protein SMU_63c can also form amyloid fibrils. P1, WapA and SMU_63c were found to significantly influence biofilm development and architecture, and all three proteins were shown by immunogold electron microscopy to reside within the fibrillar extracellular matrix of the biofilms. We also showed that SMU_63c functions as a negative regulator of biofilm cell density and genetic competence. In addition, the naturally occurring C-terminal cleavage product of P1, C123 (also known as AgII), was shown to represent the amyloidogenic moiety of this protein. Thus, P1 and WapA both represent sortase substrates that are processed to amyloidogenic truncation derivatives. Our current results suggest a novel mechanism by which certain cell surface adhesins are processed and contribute to the amyloidogenic capability of S. mutans. We further demonstrate that the polyphenolic small molecules tannic acid and epigallocatechin-3-gallate, and the benzoquinone derivative AA-861, which all inhibit amyloid fibrillization of C123 and antigen A in vitro, also inhibit S. mutans biofilm formation via P1- and WapA-dependent mechanisms, indicating that these proteins serve as therapeutic targets of anti-amyloid compounds.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

