Vaccination has minimal impact on the intrahost diversity of H3N2 influenza viruses
- PMID: 28141862
- PMCID: PMC5302840
- DOI: 10.1371/journal.ppat.1006194
Vaccination has minimal impact on the intrahost diversity of H3N2 influenza viruses
Abstract
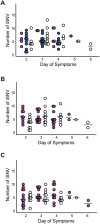
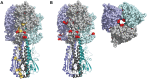
While influenza virus diversity and antigenic drift have been well characterized on a global scale, the factors that influence the virus' rapid evolution within and between human hosts are less clear. Given the modest effectiveness of seasonal vaccination, vaccine-induced antibody responses could serve as a potent selective pressure for novel influenza variants at the individual or community level. We used next generation sequencing of patient-derived viruses from a randomized, placebo-controlled trial of vaccine efficacy to characterize the diversity of influenza A virus and to define the impact of vaccine-induced immunity on within-host populations. Importantly, this study design allowed us to isolate the impact of vaccination while still studying natural infection. We used pre-season hemagglutination inhibition and neuraminidase inhibition titers to quantify vaccine-induced immunity directly and to assess its impact on intrahost populations. We identified 166 cases of H3N2 influenza over 3 seasons and 5119 person-years. We obtained whole genome sequence data for 119 samples and used a stringent and empirically validated analysis pipeline to identify intrahost single nucleotide variants at ≥1% frequency. Phylogenetic analysis of consensus hemagglutinin and neuraminidase sequences showed no stratification by pre-season HAI and NAI titer, respectively. In our study population, we found that the vast majority of intrahost single nucleotide variants were rare and that very few were found in more than one individual. Most samples had fewer than 15 single nucleotide variants across the entire genome, and the level of diversity did not significantly vary with day of sampling, vaccination status, or pre-season antibody titer. Contrary to what has been suggested in experimental systems, our data indicate that seasonal influenza vaccination has little impact on intrahost diversity in natural infection and that vaccine-induced immunity may be only a minor contributor to antigenic drift at local scales.
Conflict of interest statement
ASM has served as an ad hoc consultant to Sanofi Pasteur.
Figures



References
-
- World Health Organization. Fact sheet No 211. 2014 [Internet]. 211AD. http://www.who.int/mediacentre/factsheets/fs211/en/
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

