Expansion and stress responses of the AP2/EREBP superfamily in cotton
- PMID: 28143399
- PMCID: PMC5282909
- DOI: 10.1186/s12864-017-3517-9
Expansion and stress responses of the AP2/EREBP superfamily in cotton
Abstract
Background: The allotetraploid cotton originated from one hybridization event between an extant progenitor of Gosssypium herbaceum (A1) or G. arboreum (A2) and another progenitor, G. raimondii Ulbrich (D5) 1-1.5 million years ago (Mya). The APETALA2/ethylene-responsive element binding protein (AP2/EREBP) transcription factors constitute one of the largest and most conserved gene families in plants. They are characterized by their AP2 domain, which comprises 60-70 amino acids, and are classified into four main subfamilies: the APETALA2 (AP2), Related to ABI3/VP1 (RAV), Dehydration-Responsive Element Binding protein (DREB) and Ethylene-Responsive Factor (ERF) subfamilies. The AP2/EREBP genes play crucial roles in plant growth, development and biotic and abiotic stress responses. Hence, understanding the molecular characteristics of cotton stress tolerance and gene family expansion would undoubtedly facilitate cotton resistance breeding and evolution research.
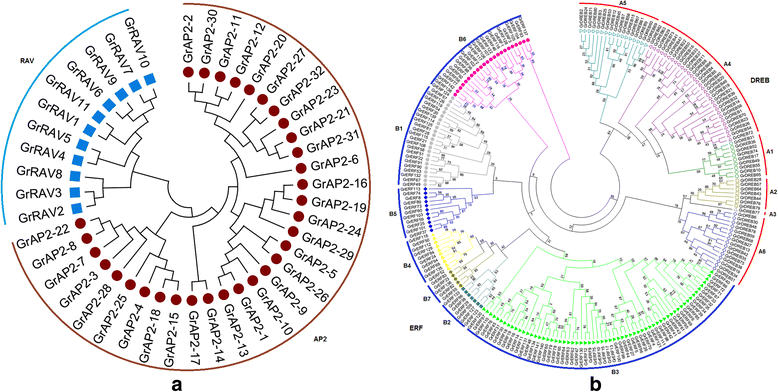
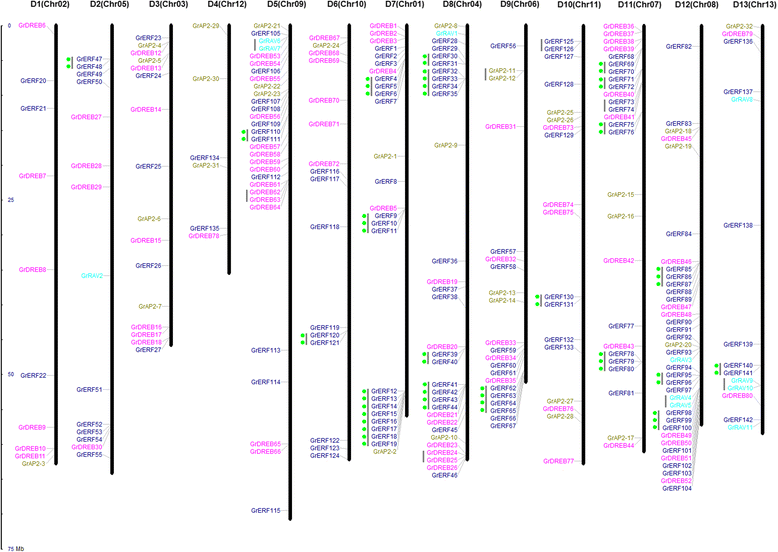
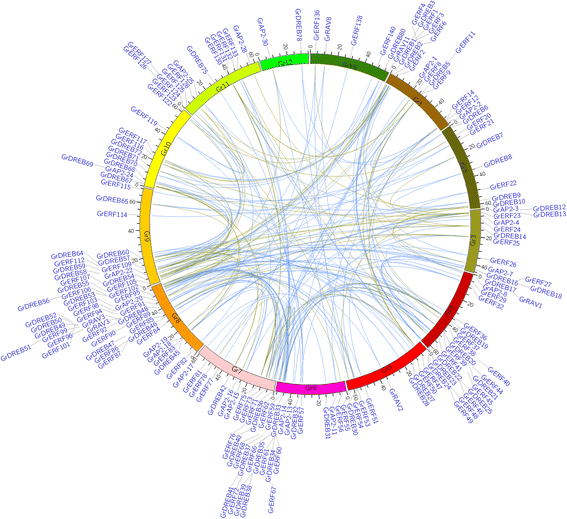
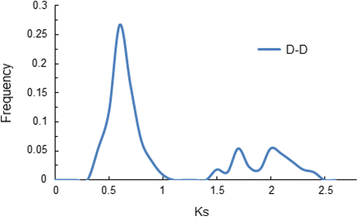
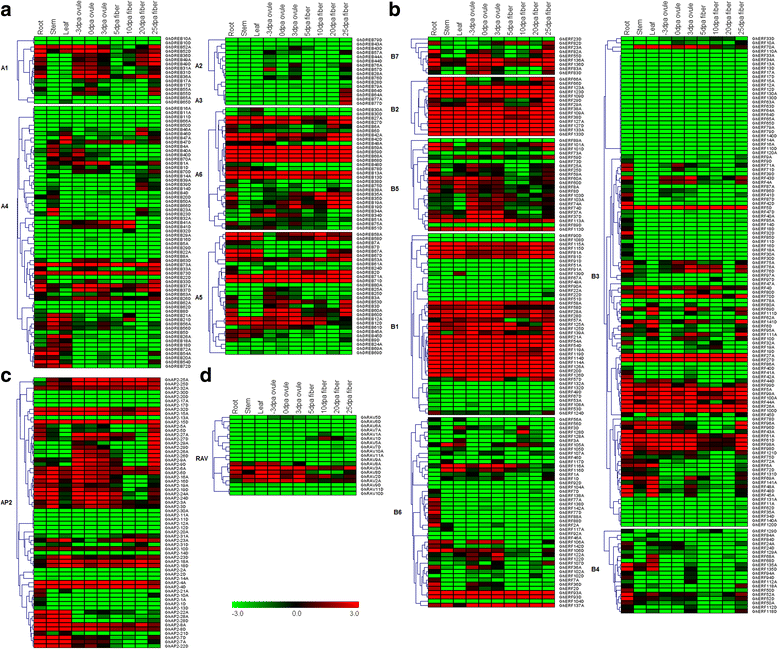
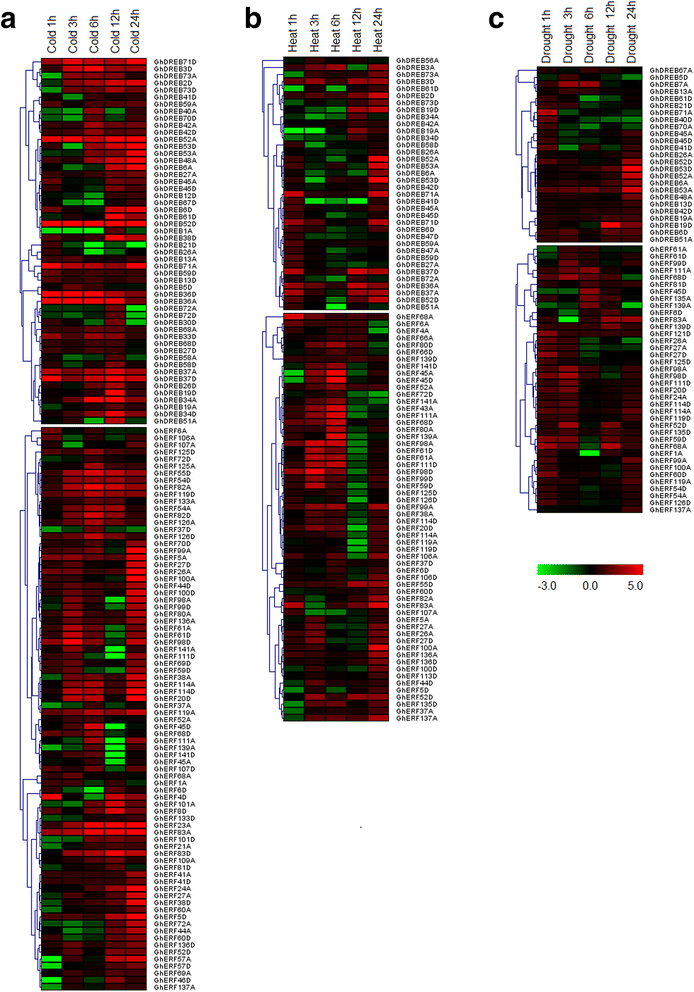
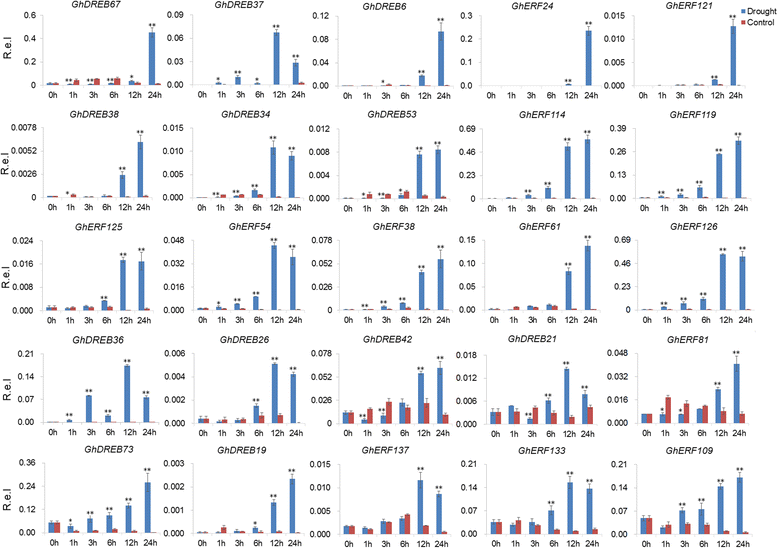
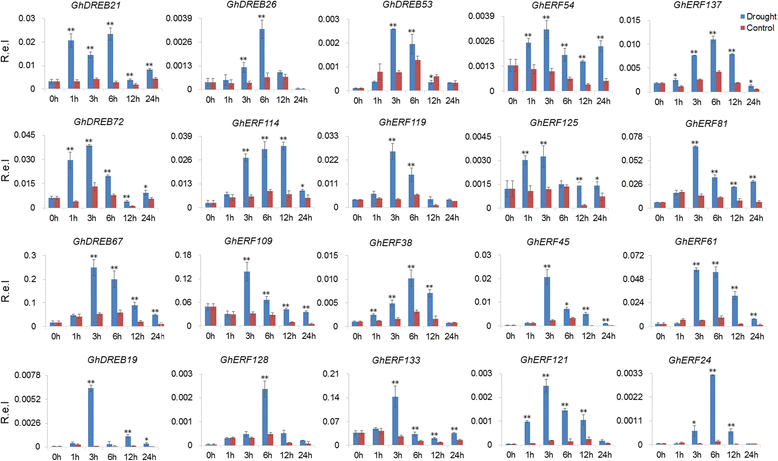
Results: A total of 269 AP2/EREBP genes were identified in the G. raimondii (D5) cotton genome. The protein domain architecture and intron/exon structure are simple and relatively conserved within each subfamily. They are distributed throughout all chromosomes but are clustered on various chromosomes due to genomic tandem duplication. We identified 73 tandem duplicated genes and 221 segmental duplicated gene pairs which contributed to the expansion of AP2/EREBP superfamily. Of them, tandem duplication was the most important force of the expansion of the B3 group. Transcriptome analysis showed that 504 AP2/EREBP genes were expressed in at least one tested G. hirsutum TM-1 tissues. In G. hirsutum, 151 non-repeated genes of the DREB and ERF subfamily genes were responsive to different stresses: 132 genes were induced by cold, 63 genes by drought and 94 genes by heat. qRT-PCR confirmed that 13 GhDREB and 15 GhERF genes were induced by cold and/or drought. No transcripts detected for 53 of the 111 tandem duplicated genes in TM-1. In addition, some homoeologous genes showed biased expression toward either A-or D-subgenome.
Conclusions: The AP2/EREBP genes were obviously expanded in Gossypium. The GhDREB and GhERF genes play crucial roles in cotton stress responses. Our genome-wide analysis of AP2/EREBP genes in cotton provides valuable information for characterizing the molecular functions of AP2/EREBP genes and reveals insights into their evolution in polyploid plants.
Keywords: Duplicated genes; Gene expansion; Gossypium; Homoeologous genes; Polyploid; Stress response.
Figures
Similar articles
-
Expansion and stress responses of AP2/EREBP superfamily in Brachypodium distachyon.Sci Rep. 2016 Feb 12;6:21623. doi: 10.1038/srep21623. Sci Rep. 2016. PMID: 26869021 Free PMC article.
-
Gene structures, classification and expression models of the AP2/EREBP transcription factor family in rice.Plant Cell Physiol. 2011 Feb;52(2):344-60. doi: 10.1093/pcp/pcq196. Epub 2010 Dec 17. Plant Cell Physiol. 2011. PMID: 21169347
-
Genome-wide comparison of AP2/ERF superfamily genes between Gossypium arboreum and G. raimondii.Genet Mol Res. 2016 Jul 29;15(3). doi: 10.4238/gmr.15038211. Genet Mol Res. 2016. PMID: 27525884
-
Advances in AP2/ERF super-family transcription factors in plant.Crit Rev Biotechnol. 2020 Sep;40(6):750-776. doi: 10.1080/07388551.2020.1768509. Epub 2020 Jun 10. Crit Rev Biotechnol. 2020. PMID: 32522044 Review.
-
The AP2/EREBP family of plant transcription factors.Biol Chem. 1998 Jun;379(6):633-46. doi: 10.1515/bchm.1998.379.6.633. Biol Chem. 1998. PMID: 9687012 Review.
Cited by
-
Morpho-Physiological and Proteomic Response of Bt-Cotton and Non-Bt Cotton to Drought Stress.Front Plant Sci. 2021 May 10;12:663576. doi: 10.3389/fpls.2021.663576. eCollection 2021. Front Plant Sci. 2021. PMID: 34040622 Free PMC article.
-
Transcriptomic responses to biotic stresses in Malus x domestica: a meta-analysis study.Sci Rep. 2018 Jan 31;8(1):1970. doi: 10.1038/s41598-018-19348-4. Sci Rep. 2018. PMID: 29386527 Free PMC article.
-
Regional Heritability Mapping of Quantitative Trait Loci Controlling Traits Related to Growth and Productivity in Popcorn (Zea mays L.).Plants (Basel). 2021 Sep 6;10(9):1845. doi: 10.3390/plants10091845. Plants (Basel). 2021. PMID: 34579378 Free PMC article.
-
A comprehensive analysis of transcriptomic data for comparison of plants with different photosynthetic pathways in response to drought stress.PLoS One. 2023 Jun 27;18(6):e0287761. doi: 10.1371/journal.pone.0287761. eCollection 2023. PLoS One. 2023. PMID: 37368898 Free PMC article.
-
Physiological and Transcriptional Analysis Reveals the Response Mechanism of Camellia vietnamensis Huang to Drought Stress.Int J Mol Sci. 2022 Oct 5;23(19):11801. doi: 10.3390/ijms231911801. Int J Mol Sci. 2022. PMID: 36233104 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

