The carotenoid biosynthetic and catabolic genes in wheat and their association with yellow pigments
- PMID: 28143400
- PMCID: PMC5286776
- DOI: 10.1186/s12864-016-3395-6
The carotenoid biosynthetic and catabolic genes in wheat and their association with yellow pigments
Abstract
Background: In plants carotenoids play an important role in the photosynthetic process and photo-oxidative protection, and are the substrate for the synthesis of abscisic acid and strigolactones. In addition to their protective role as antioxidants and precursors of vitamin A, in wheat carotenoids are important as they influence the colour (whiteness vs. yellowness) of the grain. Understanding the genetic basis of grain yellow pigments, and identifying associated markers provide the basis for improving wheat quality by molecular breeding.
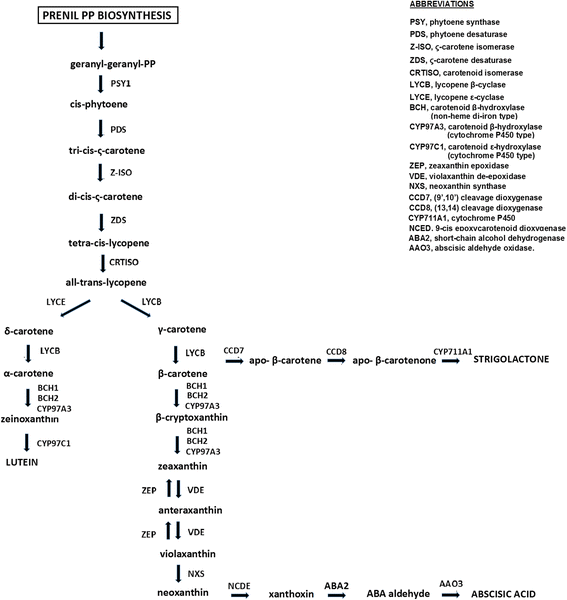
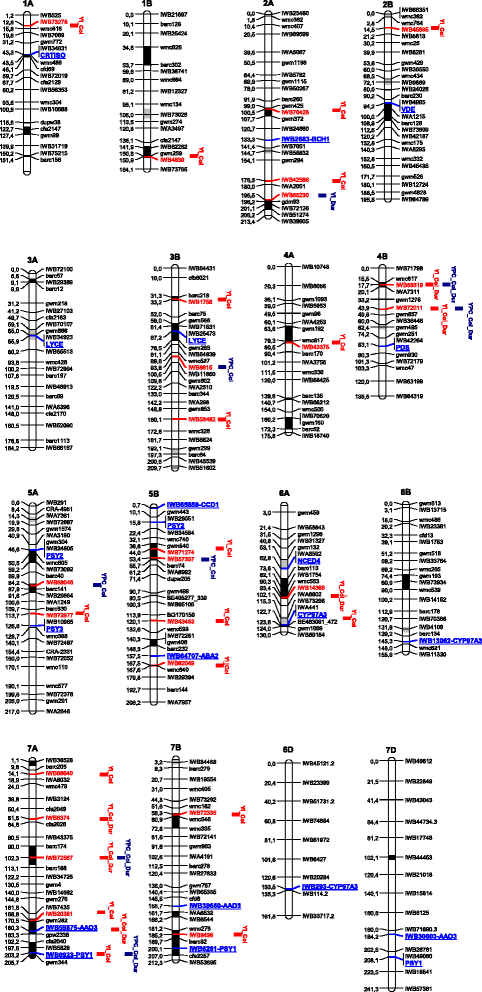
Results: Twenty-four candidate genes involved in the biosynthesis and catabolism of carotenoid compounds have been identified in wheat by comparative genomics. Single nucleotide polymorphisms (SNPs) found in the coding sequences of 19 candidate genes allowed their chromosomal location and accurate map position on two reference consensus maps to be determined. The genome-wide association study based on genotyping a tetraploid wheat collection with 81,587 gene-associated SNPs validated quantitative trait loci (QTLs) previously detected in biparental populations and discovered new QTLs for grain colour-related traits. Ten carotenoid genes mapped in chromosome regions underlying pigment content QTLs indicating possible functional relationships between candidate genes and the trait.
Conclusions: The availability of linked, candidate gene-based markers can facilitate breeding wheat cultivars with desirable levels of carotenoids. Identifying QTLs linked to carotenoid pigmentation can contribute to understanding genes underlying carotenoid accumulation in the wheat kernels. Together these outputs can be combined to exploit the genetic variability of colour-related traits for the nutritional and commercial improvement of wheat products.
Keywords: Association mapping; Carotenoids genes; Flour colour; SNP; Wheat; Yellow pigments.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

