Genomic predictions can accelerate selection for resistance against Piscirickettsia salmonis in Atlantic salmon (Salmo salar)
- PMID: 28143402
- PMCID: PMC5282740
- DOI: 10.1186/s12864-017-3487-y
Genomic predictions can accelerate selection for resistance against Piscirickettsia salmonis in Atlantic salmon (Salmo salar)
Abstract
Background: Salmon Rickettsial Syndrome (SRS) caused by Piscirickettsia salmonis is a major disease affecting the Chilean salmon industry. Genomic selection (GS) is a method wherein genome-wide markers and phenotype information of full-sibs are used to predict genomic EBV (GEBV) of selection candidates and is expected to have increased accuracy and response to selection over traditional pedigree based Best Linear Unbiased Prediction (PBLUP). Widely used GS methods such as genomic BLUP (GBLUP), SNPBLUP, Bayes C and Bayesian Lasso may perform differently with respect to accuracy of GEBV prediction. Our aim was to compare the accuracy, in terms of reliability of genome-enabled prediction, from different GS methods with PBLUP for resistance to SRS in an Atlantic salmon breeding program. Number of days to death (DAYS), binary survival status (STATUS) phenotypes, and 50 K SNP array genotypes were obtained from 2601 smolts challenged with P. salmonis. The reliability of different GS methods at different SNP densities with and without pedigree were compared to PBLUP using a five-fold cross validation scheme.
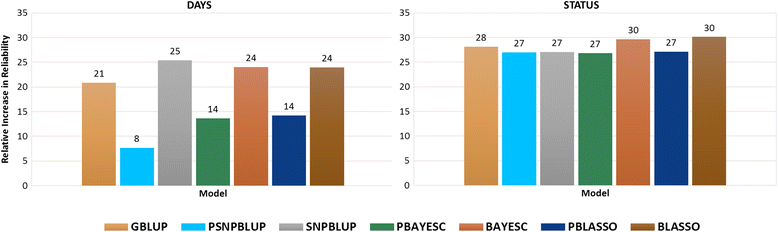
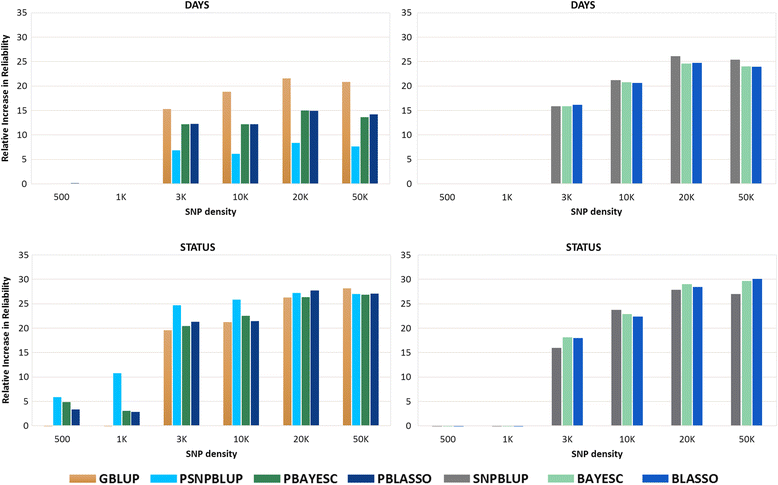
Results: Heritability estimated from GS methods was significantly higher than PBLUP. Pearson's correlation between predicted GEBV from PBLUP and GS models ranged from 0.79 to 0.91 and 0.79-0.95 for DAYS and STATUS, respectively. The relative increase in reliability from different GS methods for DAYS and STATUS with 50 K SNP ranged from 8 to 25% and 27-30%, respectively. All GS methods outperformed PBLUP at all marker densities. DAYS and STATUS showed superior reliability over PBLUP even at the lowest marker density of 3 K and 500 SNP, respectively. 20 K SNP showed close to maximal reliability for both traits with little improvement using higher densities.
Conclusions: These results indicate that genomic predictions can accelerate genetic progress for SRS resistance in Atlantic salmon and implementation of this approach will contribute to the control of SRS in Chile. We recommend GBLUP for routine GS evaluation because this method is computationally faster and the results are very similar with other GS methods. The use of lower density SNP or the combination of low density SNP and an imputation strategy may help to reduce genotyping costs without compromising gain in reliability.
Keywords: Disease resistance; Genomic selection; Reliability; Salmon Rickettsial Syndrome.
Figures
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

