In Situ Tagged nsp15 Reveals Interactions with Coronavirus Replication/Transcription Complex-Associated Proteins
- PMID: 28143984
- PMCID: PMC5285509
- DOI: 10.1128/mBio.02320-16
In Situ Tagged nsp15 Reveals Interactions with Coronavirus Replication/Transcription Complex-Associated Proteins
Abstract
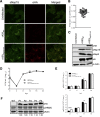
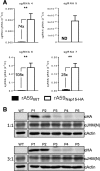
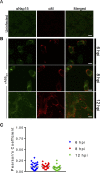
Coronavirus (CoV) replication and transcription are carried out in close proximity to restructured endoplasmic reticulum (ER) membranes in replication/transcription complexes (RTC). Many of the CoV nonstructural proteins (nsps) are required for RTC function; however, not all of their functions are known. nsp15 contains an endoribonuclease domain that is conserved in the CoV family. While the enzymatic activity and crystal structure of nsp15 are well defined, its role in replication remains elusive. nsp15 localizes to sites of RNA replication, but whether it acts independently or requires additional interactions for its function remains unknown. To begin to address these questions, we created an in situ tagged form of nsp15 using the prototypic CoV, mouse hepatitis virus (MHV). In MHV, nsp15 contains the genomic RNA packaging signal (P/S), a 95-bp RNA stem-loop structure that is not required for viral replication or nsp15 function. Utilizing this knowledge, we constructed an internal hemagglutinin (HA) tag that replaced the P/S. We found that nsp15-HA was localized to discrete perinuclear puncta and strongly colocalized with nsp8 and nsp12, both well-defined members of the RTC, but not the membrane (M) protein, involved in virus assembly. Finally, we found that nsp15 interacted with RTC-associated proteins nsp8 and nsp12 during infection, and this interaction was RNA independent. From this, we conclude that nsp15 localizes and interacts with CoV proteins in the RTC, suggesting it plays a direct or indirect role in virus replication. Furthermore, the use of in situ epitope tags could be used to determine novel nsp-nsp interactions in coronaviruses.
Importance: Despite structural and biochemical data demonstrating that the coronavirus nsp15 protein contains an endoribonuclease domain, its precise function during coronavirus infection remains unknown. In this work, we created a novel in situ tagged form of nsp15 to study interactions and localization during infection. This in situ tag was tolerated by MHV and did not affect viral replication. Utilizing this tag, we established that nsp15 localized to sites of replication but not sites of assembly throughout infection. Furthermore, we found that nsp15 interacted with the putative viral primase nsp8 and polymerase nsp12 during CoV infection. The strong association of nsp15 with replication complexes and interactions with replicative CoV enzymes suggest nsp15 is involved in CoV replication. These data and tools developed in this study help elucidate the function of nsp15 during infection and may be used to uncover other novel viral protein interactions.
Copyright © 2017 Athmer et al.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

