Osa-miR169 Negatively Regulates Rice Immunity against the Blast Fungus Magnaporthe oryzae
- PMID: 28144248
- PMCID: PMC5239796
- DOI: 10.3389/fpls.2017.00002
Osa-miR169 Negatively Regulates Rice Immunity against the Blast Fungus Magnaporthe oryzae
Abstract
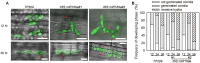
miR169 is a conserved microRNA (miRNA) family involved in plant development and stress-induced responses. However, how miR169 functions in rice immunity remains unclear. Here, we show that miR169 acts as a negative regulator in rice immunity against the blast fungus Magnaporthe oryzae by repressing the expression of nuclear factor Y-A (NF-YA) genes. The accumulation of miR169 was significantly increased in a susceptible accession but slightly fluctuated in a resistant accession upon M. oryzae infection. Consistently, the transgenic lines overexpressing miR169a became hyper-susceptible to different M. oryzae strains associated with reduced expression of defense-related genes and lack of hydrogen peroxide accumulation at the infection site. Consequently, the expression of its target genes, the NF-YA family members, was down-regulated by the overexpression of miR169a at either transcriptional or translational level. On the contrary, overexpression of a target mimicry that acts as a sponge to trap miR169a led to enhanced resistance to M. oryzae. In addition, three of miR169's target genes were also differentially up-regulated in the resistant accession upon M. oryzae infection. Taken together, our data indicate that miR169 negatively regulates rice immunity against M. oryzae by differentially repressing its target genes and provide the potential to engineer rice blast resistance via a miRNA.
Keywords: miR169; microRNA; nuclear factor Y-A; rice blast; target mimicry.
Figures






References
-
- Bonman J. M., Dios T. I. V. D., Khin M. M. (1986). Physiologic specialization of Pyricularia oryzae in the Philippines. Plant Dis. 70 767–769. 10.1094/PD-70-767 - DOI
-
- Campo S., Peris-Peris C., Sire C., Moreno A. B., Donaire L., Zytnicki M., et al. (2013). Identification of a novel microRNA (miRNA) from rice that targets an alternatively spliced transcript of the Nramp6 (Natural resistance-associated macrophage protein 6) gene involved in pathogen resistance. New Phytol. 199 212–217. 10.1111/nph.12292 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

