Enzymatic synthesis and phosphorolysis of 4(2)-thioxo- and 6(5)-azapyrimidine nucleosides by E. coli nucleoside phosphorylases
- PMID: 28144328
- PMCID: PMC5238616
- DOI: 10.3762/bjoc.12.254
Enzymatic synthesis and phosphorolysis of 4(2)-thioxo- and 6(5)-azapyrimidine nucleosides by E. coli nucleoside phosphorylases
Abstract
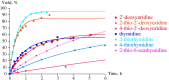
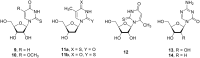
The trans-2-deoxyribosylation of 4-thiouracil (4SUra) and 2-thiouracil (2SUra), as well as 6-azauracil, 6-azathymine and 6-aza-2-thiothymine was studied using dG and E. coli purine nucleoside phosphorylase (PNP) for the in situ generation of 2-deoxy-α-D-ribofuranose-1-phosphate (dRib-1P) followed by its coupling with the bases catalyzed by either E. coli thymidine (TP) or uridine (UP) phosphorylases. 4SUra revealed satisfactory substrate activity for UP and, unexpectedly, complete inertness for TP; no formation of 2'-deoxy-2-thiouridine (2SUd) was observed under analogous reaction conditions in the presence of UP and TP. On the contrary, 2SU, 2SUd, 4STd and 2STd are good substrates for both UP and TP; moreover, 2SU, 4STd and 2'-deoxy-5-azacytidine (Decitabine) are substrates for PNP and the phosphorolysis of the latter is reversible. Condensation of 2SUra and 5-azacytosine with dRib-1P (Ba salt) catalyzed by the accordant UP and PNP in Tris∙HCl buffer gave 2SUd and 2'-deoxy-5-azacytidine in 27% and 15% yields, respectively. 6-Azauracil and 6-azathymine showed good substrate properties for both TP and UP, whereas only TP recognizes 2-thio-6-azathymine as a substrate. 5-Phenyl and 5-tert-butyl derivatives of 6-azauracil and its 2-thioxo derivative were tested as substrates for UP and TP, and only 5-phenyl- and 5-tert-butyl-6-azauracils displayed very low substrate activity. The role of structural peculiarities and electronic properties in the substrate recognition by E. coli nucleoside phosphorylases is discussed.
Keywords: 4(2)-thioxo- and 6(5)-aza-uacil and -thymine; PM3 and ab initio calculations; enzymatic glycosylation; recombinant E. coli uridine, thymidine and purine nucleoside phosphorylases; substrate properties.
Figures

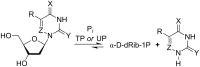



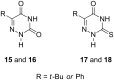

Similar articles
-
The chemoenzymatic synthesis of clofarabine and related 2'-deoxyfluoroarabinosyl nucleosides: the electronic and stereochemical factors determining substrate recognition by E. coli nucleoside phosphorylases.Beilstein J Org Chem. 2014 Jul 22;10:1657-69. doi: 10.3762/bjoc.10.173. eCollection 2014. Beilstein J Org Chem. 2014. PMID: 25161724 Free PMC article.
-
Anion exchange resins in phosphate form as versatile carriers for the reactions catalyzed by nucleoside phosphorylases.Beilstein J Org Chem. 2020 Oct 22;16:2607-2622. doi: 10.3762/bjoc.16.212. eCollection 2020. Beilstein J Org Chem. 2020. PMID: 33133292 Free PMC article.
-
Recognition of Artificial Nucleobases by E. coli Purine Nucleoside Phosphorylase versus its Ser90Ala Mutant in the Synthesis of Base-Modified Nucleosides.Chemistry. 2015 Sep 14;21(38):13401-19. doi: 10.1002/chem.201501334. Epub 2015 Jul 31. Chemistry. 2015. PMID: 26230190
-
Strained Conformations of Nucleosides in Active Sites of Nucleoside Phosphorylases.Biomolecules. 2020 Apr 5;10(4):552. doi: 10.3390/biom10040552. Biomolecules. 2020. PMID: 32260512 Free PMC article. Review.
-
Structural analyses reveal two distinct families of nucleoside phosphorylases.Biochem J. 2002 Jan 1;361(Pt 1):1-25. doi: 10.1042/0264-6021:3610001. Biochem J. 2002. PMID: 11743878 Free PMC article. Review.
Cited by
-
Strategies toward protecting group-free glycosylation through selective activation of the anomeric center.Beilstein J Org Chem. 2017 Jun 27;13:1239-1279. doi: 10.3762/bjoc.13.123. eCollection 2017. Beilstein J Org Chem. 2017. PMID: 28694870 Free PMC article. Review.
References
-
- Friedkin M, Kalckar H M. Nucleoside phosphorylases. In: Boyer P D, Lardy H, Myrbäck K, editors. The Enzymes. 2nd ed. Vol. 5. New York, USA: Academic Press; 1961. pp. 237–255.
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
