The sRNAome mining revealed existence of unique signature small RNAs derived from 5.8SrRNA from Piper nigrum and other plant lineages
- PMID: 28145468
- PMCID: PMC5286533
- DOI: 10.1038/srep41052
The sRNAome mining revealed existence of unique signature small RNAs derived from 5.8SrRNA from Piper nigrum and other plant lineages
Abstract
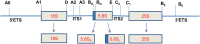
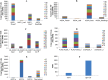
Small RNAs derived from ribosomal RNAs (srRNAs) are rarely explored in the high-throughput data of plant systems. Here, we analyzed srRNAs from the deep-sequenced small RNA libraries of Piper nigrum, a unique magnoliid plant. The 5' end of the putative long form of 5.8S rRNA (5.8SLrRNA) was identified as the site for biogenesis of highly abundant srRNAs that are unique among the Piperaceae family of plants. A subsequent comparative analysis of the ninety-seven sRNAomes of diverse plants successfully uncovered the abundant existence and precise cleavage of unique rRF signature small RNAs upstream of a novel 5' consensus sequence of the 5.8S rRNA. The major cleavage process mapped identically among the different tissues of the same plant. The differential expression and cleavage of 5'5.8S srRNAs in Phytophthora capsici infected P. nigrum tissues indicated the critical biological functions of these srRNAs during stress response. The non-canonical short hairpin precursor structure, the association with Argonaute proteins, and the potential targets of 5'5.8S srRNAs reinforced their regulatory role in the RNAi pathway in plants. In addition, this novel lineage specific small RNAs may have tremendous biological potential in the taxonomic profiling of plants.
Conflict of interest statement
The authors declare no competing financial interests.
Figures










Similar articles
-
Identifying small RNAs derived from maternal- and somatic-type rRNAs in zebrafish development.Genome. 2018 May;61(5):371-378. doi: 10.1139/gen-2017-0202. Epub 2018 Feb 9. Genome. 2018. PMID: 29425468
-
A Genome-Wide Analysis of Pathogenesis-Related Protein-1 (PR-1) Genes from Piper nigrum Reveals Its Critical Role during Phytophthora capsici Infection.Genes (Basel). 2021 Jun 30;12(7):1007. doi: 10.3390/genes12071007. Genes (Basel). 2021. PMID: 34208836 Free PMC article.
-
Unravelling the complexity of microRNA-mediated gene regulation in black pepper (Piper nigrum L.) using high-throughput small RNA profiling.Plant Cell Rep. 2016 Jan;35(1):53-63. doi: 10.1007/s00299-015-1866-x. Epub 2015 Sep 23. Plant Cell Rep. 2016. PMID: 26400683
-
Small nucleolar RNAs and pre-rRNA processing in plants.Plant Cell. 1998 May;10(5):649-57. doi: 10.1105/tpc.10.5.649. Plant Cell. 1998. PMID: 9596627 Free PMC article. Review. No abstract available.
-
RNAi in Plants: An Argonaute-Centered View.Plant Cell. 2016 Feb;28(2):272-85. doi: 10.1105/tpc.15.00920. Epub 2016 Feb 11. Plant Cell. 2016. PMID: 26869699 Free PMC article. Review.
Cited by
-
A New Specific and Sensitive RT-qPCR Method Based on Splinted 5' Ligation for the Quantitative Detection of RNA Species Shorter than microRNAs.Noncoding RNA. 2021 Sep 18;7(3):59. doi: 10.3390/ncrna7030059. Noncoding RNA. 2021. PMID: 34564321 Free PMC article.
-
Site-specific analysis reveals candidate cross-kingdom small RNAs, tRNA and rRNA fragments, and signs of fungal RNA phasing in the barley-powdery mildew interaction.Mol Plant Pathol. 2023 Jun;24(6):570-587. doi: 10.1111/mpp.13324. Epub 2023 Mar 14. Mol Plant Pathol. 2023. PMID: 36917011 Free PMC article.
-
Emerging roles of novel small non-coding regulatory RNAs in immunity and cancer.RNA Biol. 2020 Aug;17(8):1196-1213. doi: 10.1080/15476286.2020.1737442. Epub 2020 Mar 18. RNA Biol. 2020. PMID: 32186461 Free PMC article.
-
Extracellular RNA in viral-host interactions: Thinking outside the cell.Wiley Interdiscip Rev RNA. 2019 Jul;10(4):e1535. doi: 10.1002/wrna.1535. Epub 2019 Apr 8. Wiley Interdiscip Rev RNA. 2019. PMID: 30963709 Free PMC article. Review.
-
Small Non-Coding RNAs Derived From Eukaryotic Ribosomal RNA.Noncoding RNA. 2019 Feb 4;5(1):16. doi: 10.3390/ncrna5010016. Noncoding RNA. 2019. PMID: 30720712 Free PMC article. Review.
References
-
- Chen C. J. et al.. Genome-wide discovery and analysis of microRNAs and other small RNAs from rice embryogenic callus. RNA Biol. 8, 538–547 (2011). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

