Next-Generation Sequencing in Oncology: Genetic Diagnosis, Risk Prediction and Cancer Classification
- PMID: 28146134
- PMCID: PMC5343844
- DOI: 10.3390/ijms18020308
Next-Generation Sequencing in Oncology: Genetic Diagnosis, Risk Prediction and Cancer Classification
Abstract
Next-generation sequencing (NGS) technology has expanded in the last decades with significant improvements in the reliability, sequencing chemistry, pipeline analyses, data interpretation and costs. Such advances make the use of NGS feasible in clinical practice today. This review describes the recent technological developments in NGS applied to the field of oncology. A number of clinical applications are reviewed, i.e., mutation detection in inherited cancer syndromes based on DNA-sequencing, detection of spliceogenic variants based on RNA-sequencing, DNA-sequencing to identify risk modifiers and application for pre-implantation genetic diagnosis, cancer somatic mutation analysis, pharmacogenetics and liquid biopsy. Conclusive remarks, clinical limitations, implications and ethical considerations that relate to the different applications are provided.
Keywords: cancer somatic mutation; diagnostics; gene-panel; genetic modifiers; inherited cancer syndrome; next-generation sequencing; theranostics; whole-exome-sequencing; whole-genome-sequencing.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

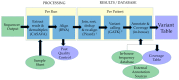
References
-
- Next-Gen-Field-Guid. [(accessed on 9 January 2016)]. Available online: http://www.molecularecologist.com/next-gen-fieldguide-2014.
-
- Illumina®. [(accessed on 10 January 2017)]. Available online: http://www.illumina.com.
-
- Ion-Torrent®. [(accessed on 7 November 2016)]. Available online: http://www.thermofisher.com.
-
- PacBio®. [(accessed on 11 November 2016)]. Available online: http://www.pacb.com/
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

