Molecular mechanism for the regulation of yeast separase by securin
- PMID: 28146474
- PMCID: PMC5302053
- DOI: 10.1038/nature21061
Molecular mechanism for the regulation of yeast separase by securin
Erratum in
-
Corrigendum: Molecular mechanism for the regulation of yeast separase by securin.Nature. 2017 Jul 27;547(7664):476. doi: 10.1038/nature23319. Epub 2017 Jul 12. Nature. 2017. PMID: 28700574
Abstract
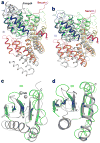
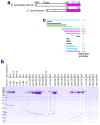
Separase is a cysteine protease with a crucial role in the dissolution of cohesion among sister chromatids during chromosome segregation. In human tumours separase is overexpressed, making it a potential target for drug discovery. The protease activity of separase is strictly regulated by the inhibitor securin, which forms a tight complex with separase and may also stabilize this enzyme. Separases are large, 140-250-kilodalton enzymes, with an amino-terminal α-helical region and a carboxy-terminal caspase-like catalytic domain. Although crystal structures of the C-terminal two domains of separase and low-resolution electron microscopy reconstructions of the separase-securin complex have been reported, the atomic structures of full-length separase and especially the complex with securin are unknown. Here we report crystal structures at up to 2.6 Å resolution of the yeast Saccharomyces cerevisiae separase-securin complex. The α-helical region of separase (also known as Esp1) contains four domains (I-IV), and a substrate-binding domain immediately precedes the catalytic domain and has tight associations with it. The separase-securin complex assumes a highly elongated structure. Residues 258-373 of securin (Pds1), named the separase interaction segment, are primarily in an extended conformation and traverse the entire length of separase, interacting with all of its domains. Most importantly, residues 258-269 of securin are located in the separase active site, illuminating the mechanism of inhibition. Biochemical studies confirm the structural observations and indicate that contacts outside the separase active site are crucial for stabilizing the complex, thereby defining an important function for the helical region of separase.
Conflict of interest statement
The authors declare no competing financial interests.
Figures





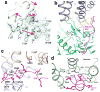





References
-
- Uhlmann F, Wernic D, Poupart MA, Koonin EV, Nasmyth K. Cleavage of cohesin by the CD clan protease separin triggers anaphase in yeast. Cell. 2000;103:375–386. - PubMed
-
- Hauf S, Waizenegger IC, Peters JM. Cohesin cleavage by separase required for anaphase and cytokinesis in human cells. Science. 2001;293:1320–1323. - PubMed
-
- Nasmyth K. Segregating sister genomes: the molecular biology of chromosome separation. Science. 2002;297:559–565. - PubMed
-
- Peters JM. The anaphase-promoting complex: proteolysis in mitosis and beyond. Mol Cell. 2002;9:931–943. - PubMed
-
- Uhlmann F. Separase regulation during mitosis. Biochem Soc Trans. 2003;70:243–251. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

