Dinucleotide Composition in Animal RNA Viruses Is Shaped More by Virus Family than by Host Species
- PMID: 28148785
- PMCID: PMC5375695
- DOI: 10.1128/JVI.02381-16
Dinucleotide Composition in Animal RNA Viruses Is Shaped More by Virus Family than by Host Species
Abstract
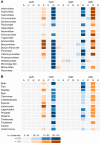
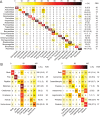
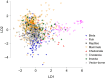
Viruses use the cellular machinery of their hosts for replication. It has therefore been proposed that the nucleotide and dinucleotide compositions of viruses should match those of their host species. If this is upheld, it may then be possible to use dinucleotide composition to predict the true host species of viruses sampled in metagenomic surveys. However, it is also clear that different taxonomic groups of viruses tend to have distinctive patterns of dinucleotide composition that may be independent of host species. To determine the relative strength of the effect of host versus virus family in shaping dinucleotide composition, we performed a comparative analysis of 20 RNA virus families from 15 host groupings, spanning two animal phyla and more than 900 virus species. In particular, we determined the odds ratios for the 16 possible dinucleotides and performed a discriminant analysis to evaluate the capability of virus dinucleotide composition to predict the correct virus family or host taxon from which it was isolated. Notably, while 81% of the data analyzed here were predicted to the correct virus family, only 62% of these data were predicted to their correct subphylum/class host and a mere 32% to their correct mammalian order. Similarly, dinucleotide composition has a weak predictive power for different hosts within individual virus families. We therefore conclude that dinucleotide composition is generally uniform within a virus family but less well reflects that of its host species. This has obvious implications for attempts to accurately predict host species from virus genome sequences alone.IMPORTANCE Determining the processes that shape virus genomes is central to understanding virus evolution and emergence. One question of particular importance is why nucleotide and dinucleotide frequencies differ so markedly between viruses. In particular, it is currently unclear whether host species or virus family has the biggest impact on dinucleotide frequencies and whether dinucleotide composition can be used to accurately predict host species. Using a comparative analysis, we show that dinucleotide composition has a strong phylogenetic association across different RNA virus families, such that dinucleotide composition can predict the family from which a virus sequence has been isolated. Conversely, dinucleotide composition has a poorer predictive power for the different host species within a virus family and across different virus families, indicating that the host has a relatively small impact on the dinucleotide composition of a virus genome.
Keywords: dinucleotide bias; evolution.
Copyright © 2017 American Society for Microbiology.
Figures

Similar articles
-
Patterns of evolution and host gene mimicry in influenza and other RNA viruses.PLoS Pathog. 2008 Jun 6;4(6):e1000079. doi: 10.1371/journal.ppat.1000079. PLoS Pathog. 2008. PMID: 18535658 Free PMC article.
-
Compositional biases and evolution of the largest plant RNA virus order Patatavirales.Int J Biol Macromol. 2023 Jun 15;240:124403. doi: 10.1016/j.ijbiomac.2023.124403. Epub 2023 Apr 18. Int J Biol Macromol. 2023. PMID: 37076075
-
Why is CpG suppressed in the genomes of virtually all small eukaryotic viruses but not in those of large eukaryotic viruses?J Virol. 1994 May;68(5):2889-97. doi: 10.1128/JVI.68.5.2889-2897.1994. J Virol. 1994. PMID: 8151759 Free PMC article.
-
Evolution and taxonomy of positive-strand RNA viruses: implications of comparative analysis of amino acid sequences.Crit Rev Biochem Mol Biol. 1993;28(5):375-430. doi: 10.3109/10409239309078440. Crit Rev Biochem Mol Biol. 1993. PMID: 8269709 Review.
-
[The great virus comeback].Biol Aujourdhui. 2013;207(3):153-68. doi: 10.1051/jbio/2013018. Epub 2013 Dec 13. Biol Aujourdhui. 2013. PMID: 24330969 Review. French.
Cited by
-
Mechanisms Underlying Host Range Variation in Flavivirus: From Empirical Knowledge to Predictive Models.J Mol Evol. 2021 Jul;89(6):329-340. doi: 10.1007/s00239-021-10013-5. Epub 2021 May 31. J Mol Evol. 2021. PMID: 34059925 Review.
-
Comparative analysis of human coronaviruses focusing on nucleotide variability and synonymous codon usage patterns.Genomics. 2021 Jul;113(4):2177-2188. doi: 10.1016/j.ygeno.2021.05.008. Epub 2021 May 19. Genomics. 2021. PMID: 34019999 Free PMC article.
-
Deciphering the SSR incidences across viral members of Coronaviridae family.Chem Biol Interact. 2020 Nov 1;331:109226. doi: 10.1016/j.cbi.2020.109226. Epub 2020 Sep 21. Chem Biol Interact. 2020. PMID: 32971122 Free PMC article.
-
Analysis of SARS-CoV-2 synonymous codon usage evolution throughout the COVID-19 pandemic.Virology. 2022 Mar;568:56-71. doi: 10.1016/j.virol.2022.01.011. Epub 2022 Feb 2. Virology. 2022. PMID: 35134624 Free PMC article.
-
Classification of COVID-19 and Other Pathogenic Sequences: A Dinucleotide Frequency and Machine Learning Approach.IEEE Access. 2020 Oct 15;8:195263-195273. doi: 10.1109/ACCESS.2020.3031387. eCollection 2020. IEEE Access. 2020. PMID: 34976561 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

