The Severe Acute Respiratory Syndrome Coronavirus Nucleocapsid Inhibits Type I Interferon Production by Interfering with TRIM25-Mediated RIG-I Ubiquitination
- PMID: 28148787
- PMCID: PMC5375661
- DOI: 10.1128/JVI.02143-16
The Severe Acute Respiratory Syndrome Coronavirus Nucleocapsid Inhibits Type I Interferon Production by Interfering with TRIM25-Mediated RIG-I Ubiquitination
Erratum in
-
Correction for Hu et al., "The Severe Acute Respiratory Syndrome Coronavirus Nucleocapsid Inhibits Type I Interferon Production by Interfering with TRIM25-Mediated RIG-I Ubiquitination".J Virol. 2020 Sep 29;94(20):e01378-20. doi: 10.1128/JVI.01378-20. Print 2020 Sep 29. J Virol. 2020. PMID: 32994231 Free PMC article. No abstract available.
Abstract
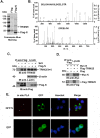
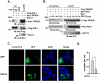
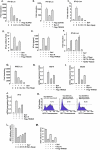
Severe acute respiratory syndrome (SARS) is a respiratory disease, caused by a coronavirus (SARS-CoV), that is characterized by atypical pneumonia. The nucleocapsid protein (N protein) of SARS-CoV plays an important role in inhibition of type I interferon (IFN) production via an unknown mechanism. In this study, the SARS-CoV N protein was found to bind to the SPRY domain of the tripartite motif protein 25 (TRIM25) E3 ubiquitin ligase, thereby interfering with the association between TRIM25 and retinoic acid-inducible gene I (RIG-I) and inhibiting TRIM25-mediated RIG-I ubiquitination and activation. Type I IFN production induced by poly I·C or Sendai virus (SeV) was suppressed by the SARS-CoV N protein. SARS-CoV replication was increased by overexpression of the full-length N protein but not N amino acids 1 to 361, which could not interact with TRIM25. These findings provide an insightful interpretation of the SARS-CoV-mediated host innate immune suppression caused by the N protein.IMPORTANCE The SARS-CoV N protein is essential for the viral life cycle and plays a key role in the virus-host interaction. We demonstrated that the interaction between the C terminus of the N protein and the SPRY domain of TRIM25 inhibited TRIM25-mediated RIG-I ubiquitination, which resulted in the inhibition of IFN production. We also found that the Middle East respiratory syndrome CoV (MERS-CoV) N protein interacted with TRIM25 and inhibited RIG-I signaling. The outcomes of these findings indicate the function of the coronavirus N protein in modulating the host's initial innate immune response.
Keywords: RIG-I; SARS coronavirus; TRIM25; interferon; nucleocapsid.
Copyright © 2017 American Society for Microbiology.
Figures





References
-
- Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, Uematsu S, Jung A, Kawai T, Ishii KJ, Yamaguchi O, Otsu K, Tsujimura T, Koh C-S, Reis e Sousa C, Matsuura Y, Fujita T, Akira S. 2006. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 441:101–105. doi: 10.1038/nature04734. - DOI - PubMed
-
- Gack MU, Albrecht RA, Urano T, Inn K-S, Huang IC, Carnero E, Farzan M, Inoue S, Jung JU, García-Sastre A. 2009. Influenza A virus NS1 targets the ubiquitin ligase TRIM25 to evade recognition by the host viral RNA sensor RIG-I. Cell Host Microbe 5:439–449. doi: 10.1016/j.chom.2009.04.006. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

