A MicroRNA Screen Identifies the Wnt Signaling Pathway as a Regulator of the Interferon Response during Flavivirus Infection
- PMID: 28148804
- PMCID: PMC5375670
- DOI: 10.1128/JVI.02388-16
A MicroRNA Screen Identifies the Wnt Signaling Pathway as a Regulator of the Interferon Response during Flavivirus Infection
Abstract
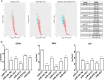
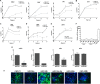
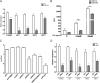
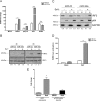
The impact of mosquito-borne flavivirus infections worldwide is significant, and many critical aspects of these viruses' biology, including virus-host interactions, host cell requirements for replication, and how virus-host interactions impact pathology, remain to be fully understood. The recent reemergence and spread of flaviviruses, including dengue virus (DENV), West Nile virus (WNV), and Zika virus (ZIKV), highlight the importance of performing basic research on this important group of pathogens. MicroRNAs (miRNAs) are small, noncoding RNAs that modulate gene expression posttranscriptionally and have been demonstrated to regulate a broad range of cellular processes. Our research is focused on identifying pro- and antiflaviviral miRNAs as a means of characterizing cellular pathways that support or limit viral replication. We have screened a library of known human miRNA mimics for their effect on the replication of three flaviviruses, DENV, WNV, and Japanese encephalitis virus (JEV), using a high-content immunofluorescence screen. Several families of miRNAs were identified as inhibiting multiple flaviviruses, including the miRNA miR-34, miR-15, and miR-517 families. Members of the miR-34 family, which have been extensively characterized for their ability to repress Wnt/β-catenin signaling, demonstrated strong antiflaviviral effects, and this inhibitory activity extended to other viruses, including ZIKV, alphaviruses, and herpesviruses. Previous research suggested a possible link between the Wnt and type I interferon (IFN) signaling pathways. Therefore, we investigated the role of type I IFN induction in the antiviral effects of the miR-34 family and confirmed that these miRNAs potentiate interferon regulatory factor 3 (IRF3) phosphorylation and translocation to the nucleus, the induction of IFN-responsive genes, and the release of type I IFN from transfected cells. We further demonstrate that the intersection between the Wnt and IFN signaling pathways occurs at the point of glycogen synthase kinase 3β (GSK3β)-TANK-binding kinase 1 (TBK1) binding, inducing TBK1 to phosphorylate IRF3 and initiate downstream IFN signaling. In this way, we have identified a novel cellular signaling network with a critical role in regulating the replication of multiple virus families. These findings highlight the opportunities for using miRNAs as tools to discover and characterize unique cellular factors involved in supporting or limiting virus replication, opening up new avenues for antiviral research.IMPORTANCE MicroRNAs are a class of small regulatory RNAs that modulate cellular processes through the posttranscriptional repression of multiple transcripts. We hypothesized that individual miRNAs may be capable of inhibiting viral replication through their effects on host proteins or pathways. To test this, we performed a high-content screen for miRNAs that inhibit the replication of three medically relevant members of the flavivirus family: West Nile virus, Japanese encephalitis virus, and dengue virus 2. The results of this screen identify multiple miRNAs that inhibit one or more of these viruses. Extensive follow-up on members of the miR-34 family of miRNAs, which are active against all three viruses as well as the closely related Zika virus, demonstrated that miR-34 functions through increasing the infected cell's ability to respond to infection through the interferon-based innate immune pathway. Our results not only add to the knowledge of how viruses interact with cellular pathways but also provide a basis for more extensive data mining by providing a comprehensive list of miRNAs capable of inhibiting flavivirus replication. Finally, the miRNAs themselves or cellular pathways identified as modulating virus infection may prove to be novel candidates for the development of therapeutic interventions.
Keywords: Wnt signaling; flavivirus; interferons; microRNA.
Copyright © 2017 American Society for Microbiology.
Figures


Similar articles
-
Cellular NONO protein binds to the flavivirus replication complex and promotes positive-strand RNA synthesis.J Virol. 2024 Dec 17;98(12):e0029724. doi: 10.1128/jvi.00297-24. Epub 2024 Nov 5. J Virol. 2024. PMID: 39499073 Free PMC article.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2021 Apr 19;4(4):CD011535. doi: 10.1002/14651858.CD011535.pub4. Cochrane Database Syst Rev. 2021. Update in: Cochrane Database Syst Rev. 2022 May 23;5:CD011535. doi: 10.1002/14651858.CD011535.pub5. PMID: 33871055 Free PMC article. Updated.
-
Behavioral interventions to reduce risk for sexual transmission of HIV among men who have sex with men.Cochrane Database Syst Rev. 2008 Jul 16;(3):CD001230. doi: 10.1002/14651858.CD001230.pub2. Cochrane Database Syst Rev. 2008. PMID: 18646068
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2017 Dec 22;12(12):CD011535. doi: 10.1002/14651858.CD011535.pub2. Cochrane Database Syst Rev. 2017. Update in: Cochrane Database Syst Rev. 2020 Jan 9;1:CD011535. doi: 10.1002/14651858.CD011535.pub3. PMID: 29271481 Free PMC article. Updated.
-
Measures implemented in the school setting to contain the COVID-19 pandemic.Cochrane Database Syst Rev. 2022 Jan 17;1(1):CD015029. doi: 10.1002/14651858.CD015029. Cochrane Database Syst Rev. 2022. Update in: Cochrane Database Syst Rev. 2024 May 2;5:CD015029. doi: 10.1002/14651858.CD015029.pub2. PMID: 35037252 Free PMC article. Updated.
Cited by
-
Hepatocellular carcinoma, hepatitis C virus infection and miRNA involvement: Perspectives for new therapeutic approaches.World J Gastroenterol. 2022 Jun 14;28(22):2417-2428. doi: 10.3748/wjg.v28.i22.2417. World J Gastroenterol. 2022. PMID: 35979260 Free PMC article. Review.
-
On the Importance of Host MicroRNAs During Viral Infection.Front Genet. 2018 Oct 2;9:439. doi: 10.3389/fgene.2018.00439. eCollection 2018. Front Genet. 2018. PMID: 30333857 Free PMC article. Review.
-
High-Throughput Fluorescence-Based Screen Identifies the Neuronal MicroRNA miR-124 as a Positive Regulator of Alphavirus Infection.J Virol. 2020 Apr 16;94(9):e02145-19. doi: 10.1128/JVI.02145-19. Print 2020 Apr 16. J Virol. 2020. PMID: 32102877 Free PMC article.
-
Advances in Zika Virus⁻Host Cell Interaction: Current Knowledge and Future Perspectives.Int J Mol Sci. 2019 Mar 4;20(5):1101. doi: 10.3390/ijms20051101. Int J Mol Sci. 2019. PMID: 30836648 Free PMC article. Review.
-
Roles of GSK-3 and β-Catenin in Antiviral Innate Immune Sensing of Nucleic Acids.Cells. 2020 Apr 7;9(4):897. doi: 10.3390/cells9040897. Cells. 2020. PMID: 32272583 Free PMC article. Review.
References
-
- Special Programme for Research & Training in Tropical Diseases (TDR). 2007. Report of Dengue Scientific Working Group. World Health Organization, Geneva, Switzerland.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

