Marine Bivalve Mollusks As Possible Indicators of Multidrug-Resistant Escherichia coli and Other Species of the Enterobacteriaceae Family
- PMID: 28149295
- PMCID: PMC5241299
- DOI: 10.3389/fmicb.2017.00024
Marine Bivalve Mollusks As Possible Indicators of Multidrug-Resistant Escherichia coli and Other Species of the Enterobacteriaceae Family
Abstract
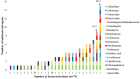
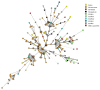
The mechanisms for the development and spread of antibacterial resistance (ABR) in bacteria residing in environmental compartments, including the marine environment, are far from understood. The objective of this study was to examine the ABR rates in Escherichia coli and other Enterobacteriaceae isolates obtained from marine bivalve mollusks collected along the Norwegian coast during a period from October 2014 to November 2015. A total of 549 bivalve samples were examined by a five times three tube most probable number method for enumeration of E. coli in bivalves resulting in 199 isolates from the positive samples. These isolates were identified by biochemical reactions and matrix Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry, showing that 90% were E. coli, while the remaining were species within the genera Klebsiella, Citrobacter, and Enterobacter. All 199 isolates recovered were susceptibility tested following the European Committee on Antimicrobial Susceptibility Testing disk diffusion method. In total, 75 of 199 (38%) isolates showed resistance to at least one antibacterial agent, while multidrug-resistance were seen in 9 (5%) isolates. One isolate conferred resistance toward 15 antibacterial agents. Among the 75 resistant isolates, resistance toward extended-spectrum penicillins (83%), aminoglycosides (16%), trimethoprim (13%), sulfonamides (11%), tetracyclines (8%), third-generation cephalosporins (7%), amphenicols (5%), nitrofurans (5%), and quinolones (5%), were observed. Whole-genome sequencing on a selection of 10 E. coli isolates identified the genes responsible for resistance, including blaCTX-M genes. To indicate the potential for horizontal gene transfer, conjugation experiments were performed on the same selected isolates. Conjugative transfer of resistance was observed for six of the 10 E. coli isolates. In order to compare E. coli isolates from bivalves with clinical strains, multiple-locus variable number tandem repeats analysis (MLVA) was applied on a selection of 30 resistant E. coli isolates. The MLVA-profiles were associated with community-acquired E. coli strains causing bacteremia. Our study indicates that bivalves represent an important tool for monitoring antibacterial resistant E. coli and other members of the Enterobacteriaceae family in the coastal environment.
Keywords: Enterobacteriaceae; Escherichia coli; antibacterial resistance; bivalve mollusks; horizontal gene transfer.
Figures
References
-
- 854/2004/EC (2004). Regulation (EC) No. 854/2004 of the European Parliament and of the Council laying down specific rules for the organisation of official controls on products of animal origin intended for human consumption. Official J. Eur. Union L 139 854/2004 83–127.
-
- Alves M. S., Pereira A., Araújo S. M., Castro B. B., Correia A., Henriques I. (2014). Seawater is a reservoir of multi-resistant Escherichia coli, including strains hosting plasmid-mediated quinolones resistance and extended-spectrum beta-lactamases genes. Front. Microbiol. 5:426 10.3389/fmicb.2014.00426 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

