An open and transparent process to select ELIXIR Node Services as implemented by ELIXIR-UK
- PMID: 28149502
- PMCID: PMC5265702
- DOI: 10.12688/f1000research.10473.2
An open and transparent process to select ELIXIR Node Services as implemented by ELIXIR-UK
Abstract
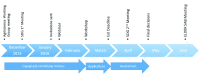
ELIXIR is the European infrastructure established specifically for the sharing and sustainability of life science data. To provide up-to-date resources and services, ELIXIR needs to undergo a continuous process of refreshing the services provided by its national Nodes. Here we present the approach taken by ELIXIR-UK to address the advice by the ELIXIR Scientific Advisory Board that Nodes need to develop " mechanisms to ensure that each Node continues to be representative of the Bioinformatics efforts within the country". ELIXIR-UK put in place an open and transparent process to identify potential ELIXIR resources within the UK during late 2015 and early to mid-2016. Areas of strategic strength were identified and Expressions of Interest in these priority areas were requested from the UK community. Criteria were established, in discussion with the ELIXIR Hub, and prospective ELIXIR-UK resources were assessed by an independent committee set up by the Node for this purpose. Of 19 resources considered, 14 were judged to be immediately ready to be included in the UK ELIXIR Node's portfolio. A further five were placed on the Node's roadmap for future consideration for inclusion. ELIXIR-UK expects to repeat this process regularly to ensure its portfolio continues to reflect its community's strengths.
Keywords: ELIXIR; ELIXIR-UK; ESFRI; e-Infrastructure.
Conflict of interest statement
Competing interests: No competing interests were disclosed.
Figures
References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

