Evolutionary origins of the emergent ST796 clone of vancomycin resistant Enterococcus faecium
- PMID: 28149688
- PMCID: PMC5267571
- DOI: 10.7717/peerj.2916
Evolutionary origins of the emergent ST796 clone of vancomycin resistant Enterococcus faecium
Abstract
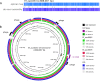
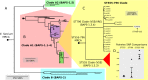
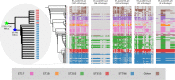
From early 2012, a novel clone of vancomycin resistant Enterococcus faecium (assigned the multi locus sequence type ST796) was simultaneously isolated from geographically separate hospitals in south eastern Australia and New Zealand. Here we describe the complete genome sequence of Ef_aus0233, a representative ST796 E. faecium isolate. We used PacBio single molecule real-time sequencing to establish a high quality, fully assembled genome comprising a circular chromosome of 2,888,087 bp and five plasmids. Comparison of Ef_aus0233 to other E. faecium genomes shows Ef_aus0233 is a member of the epidemic hospital-adapted lineage and has evolved from an ST555-like ancestral progenitor by the accumulation or modification of five mosaic plasmids and five putative prophage, acquisition of two cryptic genomic islands, accrued chromosomal single nucleotide polymorphisms and a 80 kb region of recombination, also gaining Tn1549 and Tn916, transposons conferring resistance to vancomycin and tetracycline respectively. The genomic dissection of this new clone presented here underscores the propensity of the hospital E. faecium lineage to change, presumably in response to the specific conditions of hospital and healthcare environments.
Keywords: Accessory genome; Antibiotic resistance; Comparative genomics; Enterococcus faecium; Evolution; Genome sequence; Hospital adapted; PacBio; Recombination; Vancomycin resistant.
Conflict of interest statement
The authors declare there are no competing interests.
Figures


References
-
- Carter GP, Buultjens AH, Ballard SA, Baines S, Takehiro T, Strachan J, Johnson PDR, Ferguson JK, Seemann T, Stinear TP, Howden BP. Emergence of endemic MLST non-typeable vancomycin-resistant Enterococcus faecium. Journal of Antimicrobial Chemotherapy. 2016;71:3367–3371. doi: 10.1093/jac/dkw314. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

