Bioengineering a bacterial pathogen to assemble its own particulate vaccine capable of inducing cellular immunity
- PMID: 28150705
- PMCID: PMC5288705
- DOI: 10.1038/srep41607
Bioengineering a bacterial pathogen to assemble its own particulate vaccine capable of inducing cellular immunity
Abstract
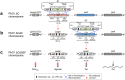
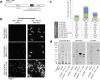
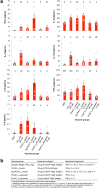
Many bacterial pathogens naturally form cellular inclusions. Here the immunogenicity of polyhydroxyalkanoate (PHA) inclusions and their use as particulate vaccines delivering a range of host derived antigens was assessed. Our study showed that PHA inclusions of pathogenic Pseudomonas aeruginosa are immunogenic mediating a specific cell-mediated immune response. Protein engineering of the PHA inclusion forming enzyme by translational fusion of epitopes from vaccine candidates outer membrane proteins OprI, OprF, and AlgE mediated self-assembly of PHA inclusions coated by these selected antigens. Mice vaccinated with isolated PHA inclusions produced a Th1 type immune response characterized by antigen-specific production of IFN-γ and IgG2c isotype antibodies. This cell-mediated immune response was found to be associated with the production of functional antibodies reacting with cells of various P. aeruginosa strains as well as facilitating opsonophagocytic killing. This study showed that cellular inclusions of pathogenic bacteria are immunogenic and can be engineered to display selected antigens suitable to serve as particulate subunit vaccines against infectious diseases.
Conflict of interest statement
B.H.A.R. is founding inventor and shareholder of PolyBatics Ltd.
Figures




References
-
- Draper J. et al. Polyhydroxyalkanoate inclusions: polymer synthesis, self-assembly and display technology. In: Bionanotechnology: biological self assembly and its applications (ed^(eds Rehm B. H. A.) Caister Academic Press (2013).
-
- Foged C. Subunit vaccines of the future: the need for safe, customized and optimized particulate delivery systems. Therapeutic delivery 2, 1057–1077 (2011). - PubMed
-
- Leleux J. & Roy K. Micro and nanoparticle‐based delivery systems for vaccine immunotherapy: an immunological and materials perspective. Advanced healthcare materials 2, 72–94 (2013). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

