Full Sequence Amino Acid Scanning of θ-Defensin RTD-1 Yields a Potent Anthrax Lethal Factor Protease Inhibitor
- PMID: 28151653
- PMCID: PMC5807008
- DOI: 10.1021/acs.jmedchem.6b01689
Full Sequence Amino Acid Scanning of θ-Defensin RTD-1 Yields a Potent Anthrax Lethal Factor Protease Inhibitor
Abstract
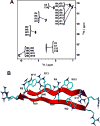
θ-Defensin RTD-1 is a noncompetitive inhibitor of anthrax lethal factor (LF) protease (IC50 = 390 ± 20 nM, Ki = 365 ± 20 nM) and a weak inhibitor of other mammalian metalloproteases such as TNFα converting enzyme (TACE) (Ki = 4.45 ± 0.48 μM). Using full sequence amino acid scanning in combination with a highly efficient "one-pot" cyclization-folding approach, we obtained an RTD-1-based peptide that was around 10 times more active than wild-type RTD-1 in inhibiting LF protease (IC50 = 43 ± 3 nM, Ki = 18 ± 1 nM). The most active peptide was completely symmetrical, rich in Arg and Trp residues, and able to adopt a native RTD-1-like structure. These results show the power of optimized chemical peptide synthesis approaches for the efficient production of libraries of disulfide-rich backbone-cyclized peptides to quickly perform structure-activity relationship studies for optimizing protease inhibitors.
Figures




References
-
- Tang YQ, Yuan J, Osapay G, Osapay K, Tran D, Miller CJ, Ouellette AJ, Selsted ME. A cyclic antimicrobial peptide produced in primate leukocytes by the ligation of two truncated alpha-defensins. Science. 1999;286:498–502. - PubMed
-
- Selsted ME, Ouellette AJ. Mammalian defensins in the antimicrobial immune response. Nat Immunol. 2005;6:551–557. - PubMed
-
- Lehrer RI. Primate defensins. Nat Rev Microbiol. 2004;2:727–738. - PubMed
-
- Rosengren KJ, Daly NL, Fornander LM, Jonsson LM, Shirafuji Y, Qu X, Vogel HJ, Ouellette AJ, Craik DJ. Structural and functional characterization of the conserved salt bridge in mammalian paneth cell alpha-defensins: solution structures of mouse CRYPTDIN-4 and (E15D)-CRYPTDIN-4. J Biol Chem. 2006;281:28068–28078. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Chemical Information
Miscellaneous

