Multiple UBXN family members inhibit retrovirus and lentivirus production and canonical NFκΒ signaling by stabilizing IκBα
- PMID: 28152074
- PMCID: PMC5308826
- DOI: 10.1371/journal.ppat.1006187
Multiple UBXN family members inhibit retrovirus and lentivirus production and canonical NFκΒ signaling by stabilizing IκBα
Abstract
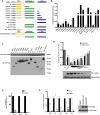
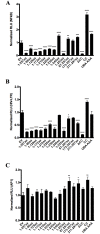
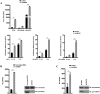
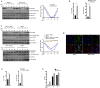
UBXN proteins likely participate in the global regulation of protein turnover, and we have shown that UBXN1 interferes with RIG-I-like receptor (RLR) signaling by interacting with MAVS and impeding its downstream effector functions. Here we demonstrate that over-expression of multiple UBXN family members decreased lentivirus and retrovirus production by several orders-of-magnitude in single cycle assays, at the level of long terminal repeat-driven transcription, and three family members, UBXN1, N9, and N11 blocked the canonical NFκB pathway by binding to Cullin1 (Cul1), inhibiting IκBα degradation. Multiple regions of UBXN1, including its UBA domain, were critical for its activity. Elimination of UBXN1 resulted in early murine embryonic lethality. shRNA-mediated knockdown of UBXN1 enhanced human immunodeficiency virus type 1 (HIV) production up to 10-fold in single cycle assays. In primary human fibroblasts, knockdown of UBXN1 caused prolonged degradation of IκBα and enhanced NFκB signaling, which was also observed after CRISPR-mediated knockout of UBXN1 in mouse embryo fibroblasts. Knockout of UBXN1 significantly up- and down-regulated hundreds of genes, notably those of several cell adhesion and immune signaling pathways. Reduction in UBXN1 gene expression in Jurkat T cells latently infected with HIV resulted in enhanced HIV gene expression, consistent with the role of UBXN1 in modulating the NFκB pathway. Based upon co-immunoprecipitation studies with host factors known to bind Cul1, models are presented as to how UBXN1 could be inhibiting Cul1 activity. The ability of UBXN1 and other family members to negatively regulate the NFκB pathway may be important for dampening the host immune response in disease processes and also re-activating quiescent HIV from latent viral reservoirs in chronically infected individuals.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
Ubiquitin-associated domain-containing ubiquitin regulatory X (UBX) protein UBXN1 is a negative regulator of nuclear factor κB (NF-κB) signaling.J Biol Chem. 2015 Apr 17;290(16):10395-405. doi: 10.1074/jbc.M114.631689. Epub 2015 Feb 13. J Biol Chem. 2015. PMID: 25681446 Free PMC article.
-
The CRISPR/Cas9 system targeting EGFR exon 17 abrogates NF-κB activation via epigenetic modulation of UBXN1 in EGFRwt/vIII glioma cells.Cancer Lett. 2017 Mar 1;388:269-280. doi: 10.1016/j.canlet.2016.12.011. Epub 2016 Dec 18. Cancer Lett. 2017. PMID: 27998759
-
Inducible expression of IkappaBalpha repressor mutants interferes with NF-kappaB activity and HIV-1 replication in Jurkat T cells.J Biol Chem. 1998 Mar 27;273(13):7431-40. doi: 10.1074/jbc.273.13.7431. J Biol Chem. 1998. PMID: 9516441
-
Cellular and viral protein interactions regulating I kappa B alpha activity during human retrovirus infection.J Leukoc Biol. 1997 Jul;62(1):82-92. doi: 10.1002/jlb.62.1.82. J Leukoc Biol. 1997. PMID: 9225998 Review.
-
Macrophage signaling in HIV-1 infection.Retrovirology. 2010 Apr 9;7:34. doi: 10.1186/1742-4690-7-34. Retrovirology. 2010. PMID: 20380698 Free PMC article. Review.
Cited by
-
UBXN1 maintains ER proteostasis and represses UPR activation by modulating translation.EMBO Rep. 2024 Feb;25(2):672-703. doi: 10.1038/s44319-023-00027-z. Epub 2024 Jan 2. EMBO Rep. 2024. PMID: 38177917 Free PMC article.
-
PTRF/Cavin-1 as a Novel RNA-Binding Protein Expedites the NF-κB/PD-L1 Axis by Stabilizing lncRNA NEAT1, Contributing to Tumorigenesis and Immune Evasion in Glioblastoma.Front Immunol. 2022 Jan 6;12:802795. doi: 10.3389/fimmu.2021.802795. eCollection 2021. Front Immunol. 2022. PMID: 35069587 Free PMC article.
-
Genetic variants at the chromosomal region 2q21.3 underlying inhibitor development in patients with severe haemophilia A.Haemophilia. 2022 Mar;28(2):270-277. doi: 10.1111/hae.14503. Epub 2022 Feb 19. Haemophilia. 2022. PMID: 35182444 Free PMC article.
-
Intrinsic cardiac adrenergic cells contribute to LPS-induced myocardial dysfunction.Commun Biol. 2022 Jan 25;5(1):96. doi: 10.1038/s42003-022-03007-6. Commun Biol. 2022. PMID: 35079095 Free PMC article.
-
DNA methylation loci in placenta associated with birthweight and expression of genes relevant for early development and adult diseases.Clin Epigenetics. 2020 Jun 3;12(1):78. doi: 10.1186/s13148-020-00873-x. Clin Epigenetics. 2020. PMID: 32493484 Free PMC article. Clinical Trial.
References
-
- Meyer H.H., Kondo H., and Warren G. The p47 co-factor regulates the ATPase activity of the membrane fusion protein, p97. FEBS Lett 1998; 437, 255–257. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

