The mutational landscape of ocular marginal zone lymphoma identifies frequent alterations in TNFAIP3 followed by mutations in TBL1XR1 and CREBBP
- PMID: 28152507
- PMCID: PMC5370020
- DOI: 10.18632/oncotarget.14928
The mutational landscape of ocular marginal zone lymphoma identifies frequent alterations in TNFAIP3 followed by mutations in TBL1XR1 and CREBBP
Erratum in
-
Correction: The mutational landscape of ocular marginal zone lymphoma identifies frequent alterations in TNFAIP3 followed by mutations in TBL1XR1 and CREBBP.Oncotarget. 2018 Aug 28;9(67):32882. doi: 10.18632/oncotarget.26080. eCollection 2018 Aug 28. Oncotarget. 2018. PMID: 30214692 Free PMC article.
Abstract
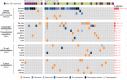
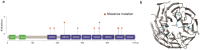
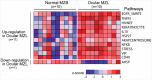
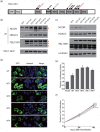
Ocular marginal zone lymphoma is a common type of low-grade B-cell lymphoma. To investigate the genomic changes that occur in ocular marginal zone lymphoma, we analyzed 10 cases of ocular marginal zone lymphoma using whole-genome and RNA sequencing and an additional 38 cases using targeted sequencing. Major genetic alterations affecting genes involved in nuclear factor (NF)-κB pathway activation (60%), chromatin modification and transcriptional regulation (44%), and B-cell differentiation (23%) were identified. In whole-genome sequencing, the 6q23.3 region containing TNFAIP3 was deleted in 5 samples (50%). In addition, 5 structural variation breakpoints in the first intron of IL20RA located in the 6q23.3 region was found in 3 samples (30%). In targeted sequencing, a disruptive mutation of TNFAIP3 was the most common alteration (54%), followed by mutations of TBL1XR1 (18%), cAMP response element binding proteins (CREBBP) (17%) and KMT2D (6%). All TBL1XR1 mutations were located within the WD40 domain, and TBL1XR1 mutants transfected into 293T cells increased TBL1XR1 binding with nuclear receptor corepressor (NCoR), leading to increased degradation of NCoR and the activation of NF-κB and JUN target genes. This study confirms genes involving in the activation of the NF-kB signaling pathway is the major driver in the oncogenesis of ocular MZL.
Keywords: RNA sequencing; marginal zone lymphoma; mutation; ocular; whole-genome sequencing.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

Similar articles
-
High throughput sequencing reveals high specificity of TNFAIP3 mutations in ocular adnexal marginal zone B-cell lymphomas.Hematol Oncol. 2020 Aug;38(3):284-292. doi: 10.1002/hon.2718. Epub 2020 Feb 16. Hematol Oncol. 2020. PMID: 32012328
-
TNFAIP3 is the target gene of chromosome band 6q23.3-q24.1 loss in ocular adnexal marginal zone B cell lymphoma.Genes Chromosomes Cancer. 2008 Jan;47(1):1-7. doi: 10.1002/gcc.20499. Genes Chromosomes Cancer. 2008. PMID: 17886247
-
Mutation analysis of NF-κB signal pathway-related genes in ocular MALT lymphoma.Int J Clin Exp Pathol. 2012;5(5):436-41. Epub 2012 May 23. Int J Clin Exp Pathol. 2012. PMID: 22808296 Free PMC article.
-
NF-κB deregulation in splenic marginal zone lymphoma.Semin Cancer Biol. 2016 Aug;39:61-7. doi: 10.1016/j.semcancer.2016.08.002. Epub 2016 Aug 5. Semin Cancer Biol. 2016. PMID: 27503810 Review.
-
MALT lymphoma: A paradigm of NF-κB dysregulation.Semin Cancer Biol. 2016 Aug;39:49-60. doi: 10.1016/j.semcancer.2016.07.003. Epub 2016 Jul 21. Semin Cancer Biol. 2016. PMID: 27452667 Review.
Cited by
-
Primary hepatopancreatobiliary lymphoma: Pathogenesis, diagnosis, and management.Front Oncol. 2022 Aug 30;12:951062. doi: 10.3389/fonc.2022.951062. eCollection 2022. Front Oncol. 2022. PMID: 36110965 Free PMC article. Review.
-
Genomic evolution of ibrutinib-resistant clones in Waldenström macroglobulinaemia.Br J Haematol. 2020 Jun;189(6):1165-1170. doi: 10.1111/bjh.16463. Epub 2020 Feb 27. Br J Haematol. 2020. PMID: 32103491 Free PMC article. Clinical Trial.
-
The art of war: using genetic insights to understand and harness radiation sensitivity in hematologic malignancies.Front Oncol. 2025 Mar 21;14:1478078. doi: 10.3389/fonc.2024.1478078. eCollection 2024. Front Oncol. 2025. PMID: 40191738 Free PMC article. Review.
-
Chromatin Regulator SPEN/SHARP in X Inactivation and Disease.Cancers (Basel). 2021 Apr 1;13(7):1665. doi: 10.3390/cancers13071665. Cancers (Basel). 2021. PMID: 33916248 Free PMC article. Review.
-
The Mutation of BTG2 Gene Predicts a Poor Outcome in Primary Testicular Diffuse Large B-Cell Lymphoma.J Inflamm Res. 2022 Mar 10;15:1757-1769. doi: 10.2147/JIR.S341355. eCollection 2022. J Inflamm Res. 2022. PMID: 35300216 Free PMC article.
References
-
- Streubel B, Simonitsch-Klupp I, Müllauer L, Lamprecht A, Huber D, Siebert R, Stolte M, Trautinger F, Lukas J, Püspök A, Formanek M, Assanasen T, Müller-Hermelink HK, et al. Variable frequencies of MALT lymphoma-associated genetic aberrations in MALT lymphomas of different sites. Leukemia. 2004(18):1722–1726. - PubMed
-
- Streubel B, Lamprecht A, Dierlamm J, Cerroni L, Stolte M, Ott G, Raderer M, Chott A. T(14;18)(q32;q21) involving IGH and MALT1 is a frequent chromosomal aberration in MALT lymphoma. Blood. 2003;101:2335–2339. - PubMed
-
- Streubel B, Vinatzer U, Lamprecht A, Raderer M, Chott A. T(3;14)(p14.1;q32) involving IGH and FOXP1 is a novel recurrent chromosomal aberration in MALT lymphoma. Leukemia. 2005;19:652–658. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

