The structure of the yeast mitochondrial ribosome
- PMID: 28154081
- PMCID: PMC5295643
- DOI: 10.1126/science.aal2415
The structure of the yeast mitochondrial ribosome
Abstract
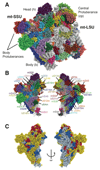
Mitochondria have specialized ribosomes (mitoribosomes) dedicated to the expression of the genetic information encoded by their genomes. Here, using electron cryomicroscopy, we have determined the structure of the 75-component yeast mitoribosome to an overall resolution of 3.3 angstroms. The mitoribosomal small subunit has been built de novo and includes 15S ribosomal RNA (rRNA) and 34 proteins, including 14 without homologs in the evolutionarily related bacterial ribosome. Yeast-specific rRNA and protein elements, including the acquisition of a putatively active enzyme, give the mitoribosome a distinct architecture compared to the mammalian mitoribosome. At an expanded messenger RNA channel exit, there is a binding platform for translational activators that regulate translation in yeast but not mammalian mitochondria. The structure provides insights into the evolution and species-specific specialization of mitochondrial translation.
Copyright © 2017, American Association for the Advancement of Science.
Figures



References
-
- Gray MW, Burger G, Lang BF. Mitochondrial evolution. Science. 1999;283:1476–1481. - PubMed
-
- Ott M, Amunts A, Brown A. Organization and Regulation of Mitochondrial Protein Synthesis. Annu Rev Biochem. 2016;85:77–101. - PubMed
-
- Greber BJ, Ban N. Structure and Function of the Mitochondrial Ribosome. Annu Rev Biochem. 2016;85:103–132. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

