Caveolae provide a specialized membrane environment for respiratory syncytial virus assembly
- PMID: 28154158
- PMCID: PMC5358342
- DOI: 10.1242/jcs.198853
Caveolae provide a specialized membrane environment for respiratory syncytial virus assembly
Abstract
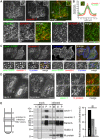
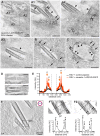
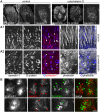
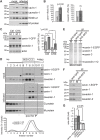
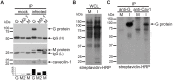
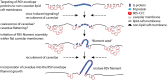
Respiratory syncytial virus (RSV) is an enveloped virus that assembles into filamentous virus particles on the surface of infected cells. Morphogenesis of RSV is dependent upon cholesterol-rich (lipid raft) membrane microdomains, but the specific role of individual raft molecules in RSV assembly is not well defined. Here, we show that RSV morphogenesis occurs within caveolar membranes and that both caveolin-1 and cavin-1 (also known as PTRF), the two major structural and functional components of caveolae, are actively recruited to and incorporated into the RSV envelope. The recruitment of caveolae occurred just prior to the initiation of RSV filament assembly, and was dependent upon an intact actin network as well as a direct physical interaction between caveolin-1 and the viral G protein. Moreover, cavin-1 protein levels were significantly increased in RSV-infected cells, leading to a virus-induced change in the stoichiometry and biophysical properties of the caveolar coat complex. Our data indicate that RSV exploits caveolae for its assembly, and we propose that the incorporation of caveolae into the virus contributes to defining the biological properties of the RSV envelope.
Keywords: Caveolae; Caveolin; Cavin; Respiratory syncytial virus; Virus assembly; Virus envelope.
© 2017. Published by The Company of Biologists Ltd.
Conflict of interest statement
The authors declare no competing or financial interests.
Figures


References
-
- Ariotti N., Fernández-Rojo M. A., Zhou Y., Hill M. M., Rodkey T. L., Inder K. L., Tanner L. B., Wenk M. R., Hancock J. F. and Parton R. G. (2014). Caveolae regulate the nanoscale organization of the plasma membrane to remotely control Ras signaling. J. Cell Biol. 204, 777-792. 10.1083/jcb.201307055 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

