A unique structural domain in Methanococcoides burtonii ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) acts as a small subunit mimic
- PMID: 28154188
- PMCID: PMC5399129
- DOI: 10.1074/jbc.M116.767145
A unique structural domain in Methanococcoides burtonii ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) acts as a small subunit mimic
Abstract
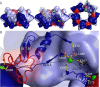
The catalytic inefficiencies of the CO2-fixing enzyme ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) often limit plant productivity. Strategies to engineer more efficient plant Rubiscos have been hampered by evolutionary constraints, prompting interest in Rubisco isoforms from non-photosynthetic organisms. The methanogenic archaeon Methanococcoides burtonii contains a Rubisco isoform that functions to scavenge the ribulose-1,5-bisphosphate (RuBP) by-product of purine/pyrimidine metabolism. The crystal structure of M. burtonii Rubisco (MbR) presented here at 2.6 Å resolution is composed of catalytic large subunits (LSu) assembled into pentamers of dimers, (L2)5, and differs from Rubiscos from higher plants where LSus are glued together by small subunits (SSu) into hexadecameric L8S8 enzymes. MbR contains a unique 29-amino acid insertion near the C terminus, which folds as a separate domain in the structure. This domain, which is visualized for the first time in this study, is located in a similar position to SSus in L8S8 enzymes between LSus of adjacent L2 dimers, where negatively charged residues coordinate around a Mg2+ ion in a fashion that suggests this domain may be important for the assembly process. The Rubisco assembly domain is thus an inbuilt SSu mimic that concentrates L2 dimers. MbR assembly is ligand-stimulated, and we show that only 6-carbon molecules with a particular stereochemistry at the C3 carbon can induce oligomerization. Based on MbR structure, subunit arrangement, sequence, phylogenetic distribution, and function, MbR and a subset of Rubiscos from the Methanosarcinales order are proposed to belong to a new Rubisco subgroup, named form IIIB.
Keywords: X-ray crystallography; archaea; carbon fixation; metal ion-protein interaction; oligomerization; protein evolution; ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco); structure-function.
© 2017 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures




Similar articles
-
Substrate-induced assembly of Methanococcoides burtonii D-ribulose-1,5-bisphosphate carboxylase/oxygenase dimers into decamers.J Biol Chem. 2009 Dec 4;284(49):33876-82. doi: 10.1074/jbc.M109.050989. Epub 2009 Oct 16. J Biol Chem. 2009. PMID: 19837658 Free PMC article.
-
Crystal structure of activated ribulose-1,5-bisphosphate carboxylase/oxygenase from green alga Chlamydomonas reinhardtii complexed with 2-carboxyarabinitol-1,5-bisphosphate.J Mol Biol. 2002 Feb 22;316(3):679-91. doi: 10.1006/jmbi.2001.5381. J Mol Biol. 2002. PMID: 11866526
-
Structure of Pisum sativum Rubisco with bound ribulose 1,5-bisphosphate.Acta Crystallogr Sect F Struct Biol Cryst Commun. 2013 Jan 1;69(Pt 1):10-4. doi: 10.1107/S1744309112047549. Epub 2012 Dec 25. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2013. PMID: 23295478 Free PMC article.
-
Role of the small subunit in ribulose-1,5-bisphosphate carboxylase/oxygenase.Arch Biochem Biophys. 2003 Jun 15;414(2):141-9. doi: 10.1016/s0003-9861(03)00171-1. Arch Biochem Biophys. 2003. PMID: 12781765 Review.
-
Biogenesis and Metabolic Maintenance of Rubisco.Annu Rev Plant Biol. 2017 Apr 28;68:29-60. doi: 10.1146/annurev-arplant-043015-111633. Epub 2017 Jan 11. Annu Rev Plant Biol. 2017. PMID: 28125284 Review.
Cited by
-
Engineering Rubisco to enhance CO2 utilization.Synth Syst Biotechnol. 2023 Dec 27;9(1):55-68. doi: 10.1016/j.synbio.2023.12.006. eCollection 2024 Mar. Synth Syst Biotechnol. 2023. PMID: 38273863 Free PMC article. Review.
-
Interspecies Comparison of Interaction Energies between Photosynthetic Protein RuBisCO and 2CABP Ligand.Int J Mol Sci. 2022 Sep 26;23(19):11347. doi: 10.3390/ijms231911347. Int J Mol Sci. 2022. PMID: 36232645 Free PMC article.
-
A peptide adhesive molded by magnesium glues Rubisco's subunits together.J Biol Chem. 2017 Apr 21;292(16):6851-6852. doi: 10.1074/jbc.H116.767145. J Biol Chem. 2017. PMID: 28432177 Free PMC article.
-
The small subunit of Rubisco and its potential as an engineering target.J Exp Bot. 2023 Jan 11;74(2):543-561. doi: 10.1093/jxb/erac309. J Exp Bot. 2023. PMID: 35849331 Free PMC article. Review.
-
Synthetic CO2-fixation enzyme cascades immobilized on self-assembled nanostructures that enhance CO2/O2 selectivity of RubisCO.Biotechnol Biofuels. 2017 Jul 6;10:175. doi: 10.1186/s13068-017-0861-6. eCollection 2017. Biotechnol Biofuels. 2017. PMID: 28694846 Free PMC article.
References
-
- Andersson I. (2008) Catalysis and regulation in Rubisco. J. Exp. Bot. 59, 1555–1568 - PubMed
-
- Long S. P., Zhu X.-G., Naidu S. L., and Ort D. R. (2006) Can improvement in photosynthesis increase crop yields? Plant Cell Environ. 29, 315–330 - PubMed
-
- Durão P., Aigner H., Nagy P., Mueller-Cajar O., Hartl F. U., and Hayer-Hartl M. (2015) Opposing effects of folding and assembly chaperones on evolvability of Rubisco. Nat. Chem. Biol. 11, 148–155 - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

